6YVR
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the peptide full agonist NTS8-13 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin crystallisation chaperone, neurotensin NTS8-13 (full agonist), nonyl beta-D-glucopyranoside | | Authors: | Deluigi, M, Merklinger, L, Hilge, M, Ernst, P, Klipp, A, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
8FMZ
 
 | | Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, MiniGq, ... | | Authors: | Krumm, B.E, Diberto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
6Z4V
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with NTS8-13 | | Descriptor: | ARG-PRO-TYR-ILE-LEU, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
6ZIN
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4S
 
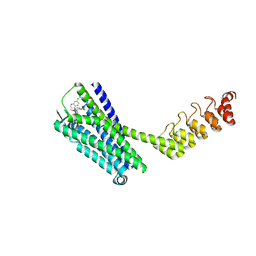 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZA8
 
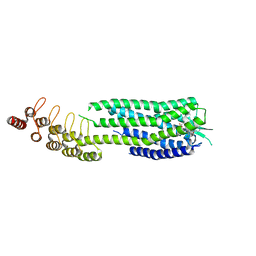 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
4BUO
 
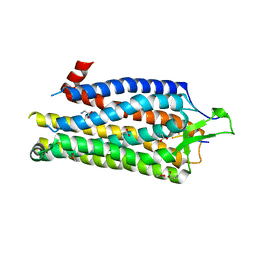 | | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4GRV
 
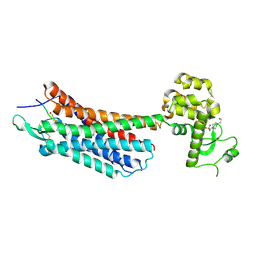 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
6Z8N
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule full agonist SRI-9829 | | Descriptor: | (2~{S})-4-methyl-2-[(1-quinolin-8-ylsulfonylindol-3-yl)carbonylamino]pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
8FN1
 
 | | CryoEM structure of Go-coupled NTSR1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Krumm, B.E, DiBerto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
8FN0
 
 | | CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, DiBerto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
4XEE
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
6Z4Q
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule inverse agonist SR142948A | | Descriptor: | 2-[[5-(2,6-dimethoxyphenyl)-1-[4-[3-(dimethylamino)propyl-methyl-carbamoyl]-2-propan-2-yl-phenyl]pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
3ZEV
 
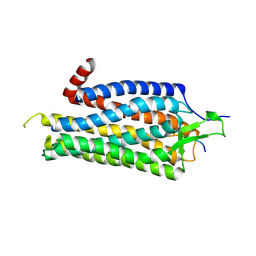 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BV0
 
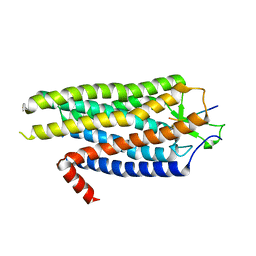 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6Z66
 
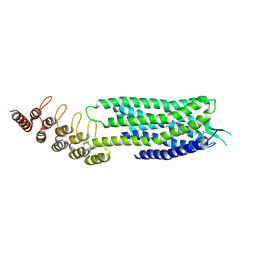 | | Crystal structure of apo-state neurotensin receptor 1 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
5T04
 
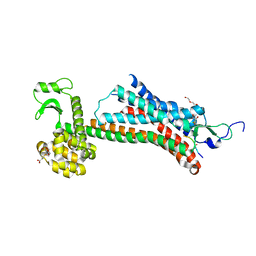 | | STRUCTURE OF CONSTITUTIVELY ACTIVE NEUROTENSIN RECEPTOR | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ARG-ARG-PRO-TYR-ILE-LEU, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B, Botos, I, Grisshammer, R. | | Deposit date: | 2016-08-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and dynamics of a constitutively active neurotensin receptor.
Sci Rep, 6, 2016
|
|
4BWB
 
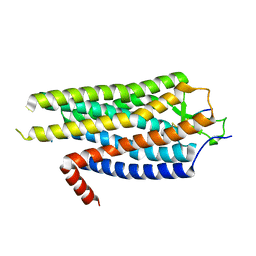 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7L0P
 
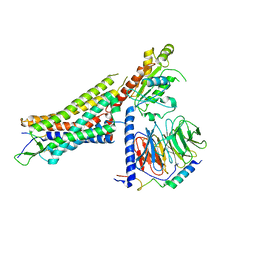 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0R
 
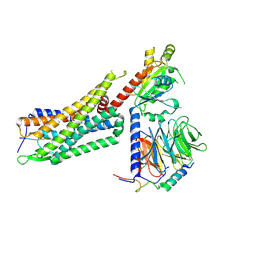 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
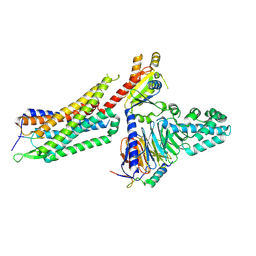 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
