3N96
 
 | |
3EHS
 
 | |
6DKS
 
 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
5EDU
 
 | |
7VN6
 
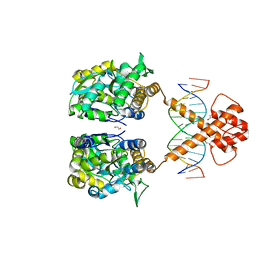 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CGCACGTGCG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*GP*CP*AP*CP*GP*TP*GP*CP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
2R6G
 
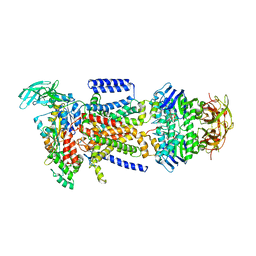 | | The Crystal Structure of the E. coli Maltose Transporter | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Maltose transport system permease protein malF, Maltose transport system permease protein malG, ... | | Authors: | Oldham, M.L, Khare, D, Quiocho, F.A, Davidson, A.L, Chen, J. | | Deposit date: | 2007-09-05 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a catalytic intermediate of the maltose transporter.
Nature, 450, 2007
|
|
8XB3
 
 | |
7XVF
 
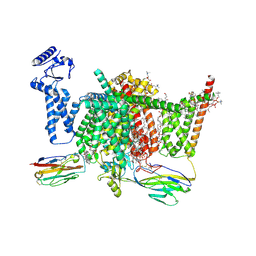 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4BLB
 
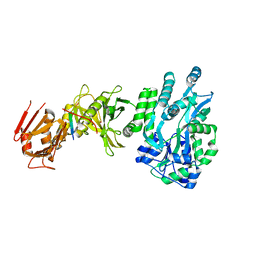 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL9
 
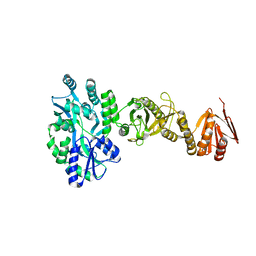 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1R6Z
 
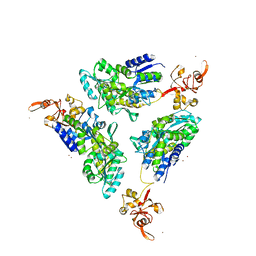 | | The Crystal Structure of the Argonaute2 PAZ domain (as a MBP fusion) | | Descriptor: | Chimera of Maltose-binding periplasmic protein and Argonaute 2, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Song, J.J, Liu, J, Tolia, N.H, Schneiderman, J, Smith, S.K, Martienssen, R.A, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2003-10-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes.
Nat.Struct.Biol., 10, 2003
|
|
7B01
 
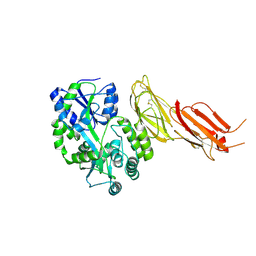 | | ADAMTS13-CUB12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein,Maltodextrin-binding protein,Maltodextrin-binding protein,ADAMTS13 CUB12,A disintegrin and metalloproteinase with thrombospondin motifs 13,A disintegrin and metalloproteinase with thrombospondin motifs 13,A disintegrin and metalloproteinase with thrombospondin motifs 13, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kim, H.J, Emsley, J. | | Deposit date: | 2020-11-18 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADAMTS13 CUB domains reveals their role in global latency.
Sci Adv, 7, 2021
|
|
6SLM
 
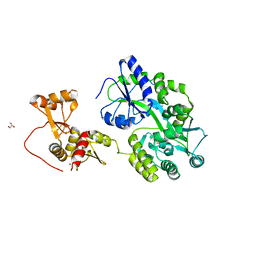 | | Crystal structure of full-length HPV31 E6 oncoprotein in complex with LXXLL peptide of ubiquitin ligase E6AP | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, ... | | Authors: | Conrady, M, Gogl, G, Cousido-Siah, A, Mitschler, A, Trave, G, Simon, C. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of High-Risk Papillomavirus 31 E6 Oncogenic Protein and Characterization of E6/E6AP/p53 Complex Formation.
J.Virol., 95, 2020
|
|
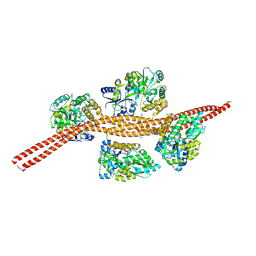
3MQ9
 
 | |
5WVN
 
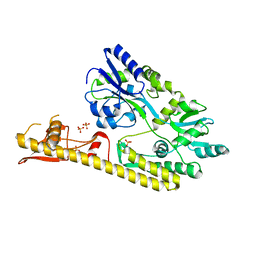 | | Crystal structure of MBS-BaeS fusion protein | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Ran, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
3RVZ
 
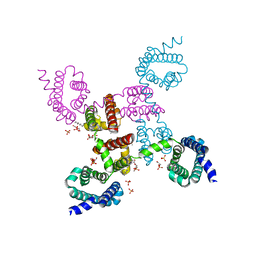 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.8 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
8G1A
 
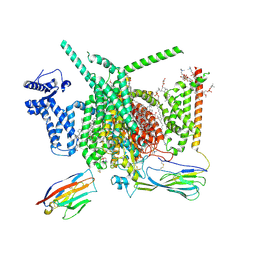 | | Cryo-EM structure of Nav1.7 with CBD | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cannabidiol inhibits Na v channels through two distinct binding sites.
Nat Commun, 14, 2023
|
|
5E7U
 
 | |
6CXS
 
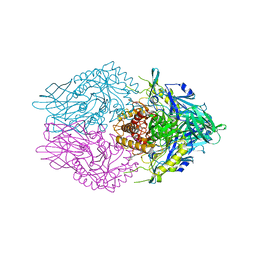 | | Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine | | Descriptor: | 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine, Beta-glucuronidase, Maltose/maltodextrin-binding periplasmic protein | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted inhibition of gut bacterial beta-glucuronidase activity enhances anticancer drug efficacy.
Proc.Natl.Acad.Sci.USA, 2020
|
|
7XQC
 
 | |
5V6Y
 
 | |
4NUF
 
 | | Crystal Structure of SHP/EID1 | | Descriptor: | EID1 peptide, Maltose ABC transporter periplasmic protein, Nuclear receptor subfamily 0 group B member 2 chimeric construct, ... | | Authors: | Zhi, X, Zhou, X.E, He, Y, Zechner, C, Suino-Powell, K.M, Kliewer, S.A, Melcher, K, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into gene repression by the orphan nuclear receptor SHP.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5YUA
 
 | | Crystal structure of voltage-gated sodium channel NavAb in high-pH condition | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHAPSO, ... | | Authors: | Irie, K, Shimomura, T, Fujiyoshi, Y. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.80085564 Å) | | Cite: | Structural insight on the voltage dependence of prokaryotic voltage gated sodium channel NavAb.
FEBS Lett., 2017
|
|
4BLD
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6KE5
 
 | | Structure of CavAb in complex with Diltiazem and Amlodipine | | Descriptor: | CALCIUM ION, Ion transport protein, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Tang, L. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for Diltiazem Block of a Voltage-Gated Ca2+Channel.
Mol.Pharmacol., 96, 2019
|
|
