1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
1N73
 
 | | Fibrin D-Dimer, Lamprey complexed with the PEPTIDE LIGAND: GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrin alpha-1 chain, ... | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-12 | | Release date: | 2003-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface
Biochemistry, 41, 2002
|
|
1N75
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N76
 
 | | CRYSTAL STRUCTURE OF HUMAN SEMINAL LACTOFERRIN AT 3.4 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN, ... | | Authors: | Kumar, J, Weber, W, Munchau, S, Yadav, S, Singh, S.B, Sarvanan, K, Paramsivam, M, Sharma, S, Kaur, P, Bhushan, A, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of human seminal lactoferrin at 3.4A resolution
Indian J.Biochem.Biophys., 40, 2003
|
|
1N77
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N78
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and glutamol-AMP. | | Descriptor: | GLUTAMOL-AMP, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N7A
 
 | | RIP-Radiation-damage Induced Phasing | | Descriptor: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | Authors: | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | Deposit date: | 2002-11-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1N7B
 
 | | RIP-Radiation-damage Induced Phasing | | Descriptor: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | Authors: | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | Deposit date: | 2002-11-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1N7D
 
 | | Extracellular domain of the LDL receptor | | Descriptor: | 12-TUNGSTOPHOSPHATE, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Rudenko, G, Henry, L, Henderson, K, Ichtchenko, K, Brown, M.S, Goldstein, J.L, Deisenhofer, J. | | Deposit date: | 2002-11-13 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the LDL receptor extracellular domain at endosomal pH
Science, 298, 2002
|
|
1N7E
 
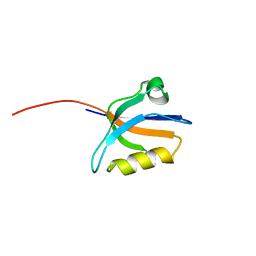 | | Crystal structure of the sixth PDZ domain of GRIP1 | | Descriptor: | AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1N7F
 
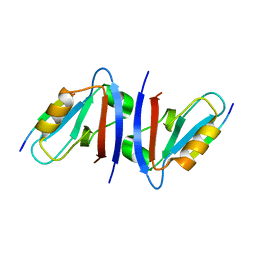 | | Crystal structure of the sixth PDZ domain of GRIP1 in complex with liprin C-terminal peptide | | Descriptor: | 8-mer peptide from interacting protein (liprin), AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1N7G
 
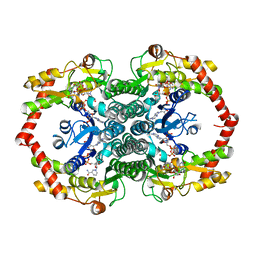 | | Crystal Structure of the GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP-rhamnose. | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE-RHAMNOSE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the MUR1 GDP-mannose 4,6-dehydratase
from A. thaliana: Implications for ligand binding and
specificity.
Biochemistry, 41, 2002
|
|
1N7H
 
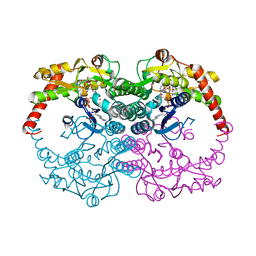 | | Crystal Structure of GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the MUR1 GDP-mannose
4,6-dehydratase from A. thaliana:
Implications for ligand binding and specificity.
Biochemistry, 41, 2002
|
|
1N7I
 
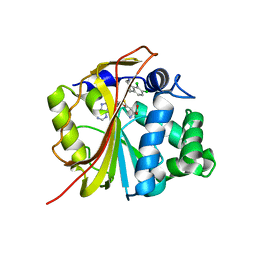 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7J
 
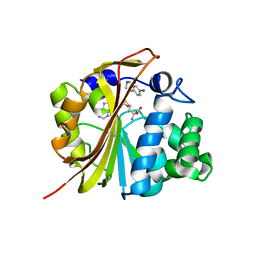 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7K
 
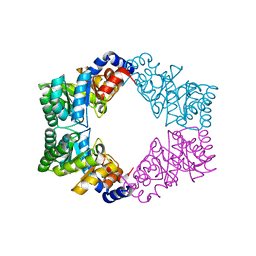 | | Unique tetrameric structure of deoxyribose phosphate aldolase from Aeropyrum pernix | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Tsuge, H, Sakuraba, H, Shimoya, I, Katunuma, N, Ago, H, Miyano, M, Ohshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-15 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The First Crystal Structure of Archaeal Aldolase. UNIQUE TETRAMERIC STRUCTURE of 2-DEOXY-D-RIBOSE-5-PHOSPHATE ALDOLASE FROM THE HYPERTHERMOPHILIC ARCHAEA Aeropyrum pernix.
J.Biol.Chem., 278, 2003
|
|
1N7L
 
 | |
1N7M
 
 | | Germline 7G12 with N-methylmesoporphyrin | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, chain H, chain L, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher IV, A.E, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-11-15 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N7N
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7O
 
 | | Streptococcus pneumoniae Hyaluronate Lyase F343V Mutant | | Descriptor: | hyaluronidase | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7P
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A/F343V Double Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7Q
 
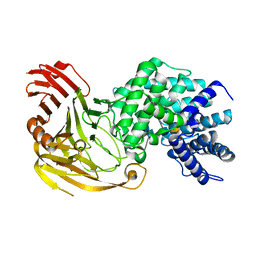 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A Double Mutant complex with hyaluronan hexasacchride | | Descriptor: | HYALURONIDASE, beta-D-galactopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7R
 
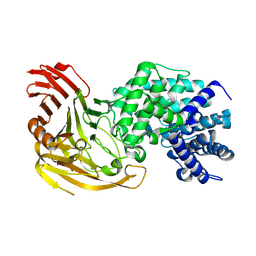 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A/F343V Mutant complex with hexasaccharide hyaluronan | | Descriptor: | HYALURONIDASE, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7S
 
 | | High Resolution Structure of a Truncated Neuronal SNARE Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, SNAP-25A, ... | | Authors: | Ernst, J.A, Brunger, A.T. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Structure, Stability, and Synaptotagmin Binding of a Truncated Neuronal SNARE Complex
J.Biol.Chem., 278, 2003
|
|
1N7T
 
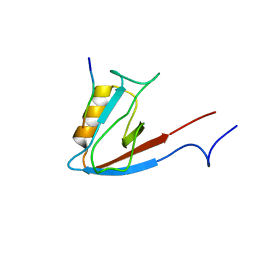 | | ERBIN PDZ domain bound to a phage-derived peptide | | Descriptor: | 99-mer peptide of densin-180-like protein, phage-derived peptide | | Authors: | Skelton, N.J, Koehler, M.F.T, Zobel, K, Wong, W.L, Yeh, S, Pisabarro, M.T, Yin, J.P, Lasky, L.A, Sidhu, S.S. | | Deposit date: | 2002-11-16 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain.
J.Biol.Chem., 278, 2003
|
|
