1JON
 
 | |
6QB8
 
 | | Human CCT:mLST8 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cuellar, J, Santiago, C, Ludlam, W.G, Bueno-Carrasco, M.T, Valpuesta, J.M, Willardson, B.M. | | Deposit date: | 2018-12-20 | | Release date: | 2019-07-03 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Nat Commun, 10, 2019
|
|
6TMU
 
 | | Crystal structure of the chaperonin gp146 from the bacteriophage EL 2 (Pseudomonas aeruginosa) in presence of ATP-BeFx, crystal form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bracher, A, Paul, S.S, Wang, H, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
1E0R
 
 | | Beta-apical domain of thermosome | | Descriptor: | THERMOSOME | | Authors: | Bosch, G, Baumeister, W, Essen, L.-O. | | Deposit date: | 2000-04-06 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
J.Mol.Biol., 301, 2000
|
|
8PE8
 
 | |
7AZP
 
 | |
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
1XCK
 
 | | Crystal structure of apo GroEL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 60 kDa chaperonin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartolucci, C, Lamba, D, Grazulis, S, Manakova, E, Heumann, H. | | Deposit date: | 2004-09-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of wild-type chaperonin GroEL
J.Mol.Biol., 354, 2005
|
|
4KI8
 
 | | Crystal structure of a GroEL-ADP complex in the relaxed allosteric state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Fei, X, Yang, D, LaRonde-LeBlanc, N, Lorimer, G.H. | | Deposit date: | 2013-05-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 A resolution.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HEL
 
 | |
2NWC
 
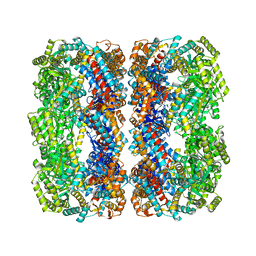 | |
4XCD
 
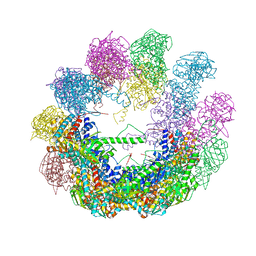 | | Crystal structure of an octadecameric TF55 complex from S. solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome subunit beta | | Authors: | Chaston, J.J, Stewart, A.G, Smits, C, Stock, D. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
1SRV
 
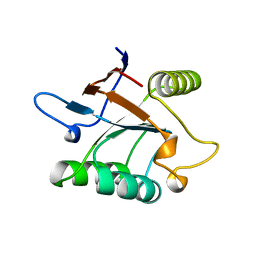 | | THERMUS THERMOPHILUS GROEL (HSP60 CLASS) FRAGMENT (APICAL DOMAIN) COMPRISING RESIDUES 192-336 | | Descriptor: | PROTEIN (GROEL (HSP60 CLASS)) | | Authors: | Walsh, M.A, Dementieva, I, Evans, G, Sanishvili, R, Joachimiak, A. | | Deposit date: | 1999-03-02 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Taking MAD to the extreme: ultrafast protein structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5CDJ
 
 | | apical domain of chloroplast chaperonin 60a | | Descriptor: | RuBisCO large subunit-binding protein subunit alpha, chloroplastic | | Authors: | Zhang, S, Yu, F, Liu, C, Gao, F. | | Deposit date: | 2015-07-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional Partition of Cpn60 alpha and Cpn60 beta Subunits in Substrate Recognition and Cooperation with Co-chaperonins
Mol Plant, 9, 2016
|
|
4XCI
 
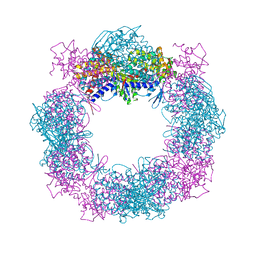 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form II | | Descriptor: | Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Chaston, J.J, Smits, C, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
2YNJ
 
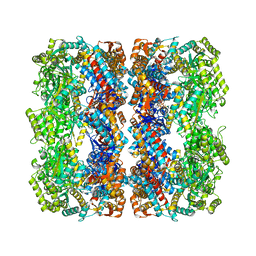 | | GroEL at sub-nanometer resolution by Constrained Single Particle Tomography | | Descriptor: | 60 KDA CHAPERONIN, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bartesaghi, A, Lecumberry, F, Sapiro, G, Subramaniam, S. | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography
Structure, 20, 2012
|
|
5GW4
 
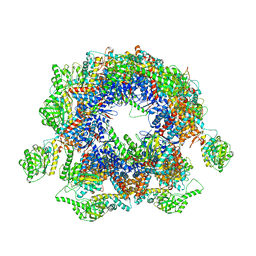 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
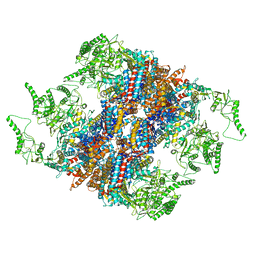 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5OPW
 
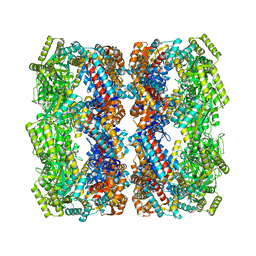 | | Crystal structure of the GroEL mutant A109C | | Descriptor: | 60 kDa chaperonin | | Authors: | Yan, X, Shi, Q, Bracher, A, Milicic, G, Singh, A.K, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | GroEL Ring Separation and Exchange in the Chaperonin Reaction.
Cell, 172, 2018
|
|
6XHJ
 
 | | Cryo-EM structure of octadecameric TF55 (beta-only) complex from S. solfataricus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Thermosome subunit beta | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural analysis of the Sulfolobus solfataricus TF55beta chaperonin by cryo-electron microscopy
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6XHI
 
 | |
6HDD
 
 | |
5DA8
 
 | | Crystal structure of chaperonin GroEL from | | Descriptor: | 60 kDa chaperonin, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Marshall, N, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chaperonin GroEL from
To Be Published
|
|
2YEY
 
 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
4WSC
 
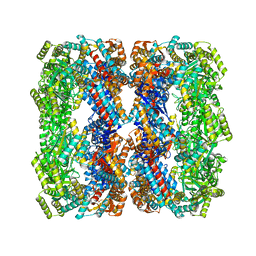 | | Crystal structure of a GroELK105A mutant | | Descriptor: | 60 kDa chaperonin | | Authors: | Lorimer, G.H, Ye, X, Fei, X, Yang, D, Corsepius, N, LaRonde, N.A. | | Deposit date: | 2014-10-26 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of a GroELK105A mutant
To Be Published
|
|
