3BPZ
 
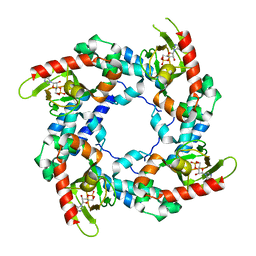 | | HCN2-I 443-460 E502K in the presence of cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Craven, K.B, Olivier, N.B, Zagotta, W.N. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | C-terminal movement during gating in cyclic nucleotide-modulated channels.
J.Biol.Chem., 283, 2008
|
|
5BV6
 
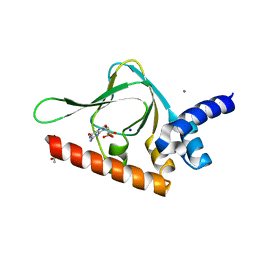 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with cGMP | | Descriptor: | ACETATE ION, CALCIUM ION, GUANOSINE-3',5'-MONOPHOSPHATE, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
4Z07
 
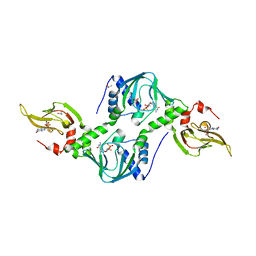 | | Co-crystal structure of the tandem CNB (CNB-A/B) domains of human PKG I beta with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kim, J.J, Reger, A.S, Arold, S.T, Kim, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PKG I:cGMP Complex Reveals a cGMP-Mediated Dimeric Interface that Facilitates cGMP-Induced Activation.
Structure, 24, 2016
|
|
6NP6
 
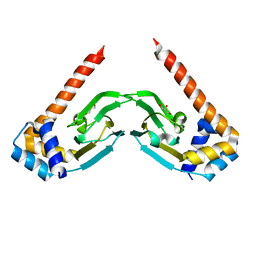 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5T3N
 
 | | Sp-2Cl-cAMPS bound to PKAR CBD2 | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-(6-amino-2-chloro-9H-purin-9-yl)-7-hydroxy-2-sulfanyltetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-2-one, IODIDE ION, cAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P. | | Deposit date: | 2016-08-26 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
5J3U
 
 | | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, Protein Kinase A | | Authors: | El Bakkouri, M, Walker, J.R, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lin, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP
To Be Published
|
|
5J48
 
 | | PKG I's Carboyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-3,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
5JD7
 
 | | PKG I's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with PET-cGMP | | Descriptor: | 3-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-6-phenyl-3,4-dihydro-9H-imidazo[1,2-a]purin-9-one, cGMP-dependent protein kinase 1 | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-04-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
2D93
 
 | |
1VP6
 
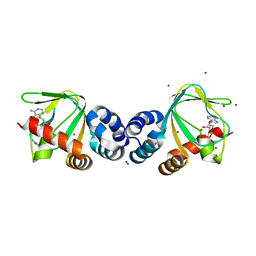 | | M.loti ion channel cylic nucleotide binding domain | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, BROMIDE ION, Cyclic-nucleotide binding domain of mesorhizobium loti CNG potassium channel | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-10-14 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel.
Cell(Cambridge,Mass.), 119, 2004
|
|
1CX4
 
 | |
5H5O
 
 | |
6SYG
 
 | |
4QX5
 
 | | Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: Insights for PKG agonist design | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Huang, G.Y, Gerlits, O.O, Blakeley, M.P, Sankaran, B, Kovalevsky, A.Y, Kim, C. | | Deposit date: | 2014-07-18 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
4QXK
 
 | | Joint X-ray/neutron structure of PKGIbeta in complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, SODIUM ION, cGMP-dependent protein kinase 1 | | Authors: | Kim, C, Gerlits, O, Kovalevsky, A, Huang, G.Y. | | Deposit date: | 2014-07-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
1RL3
 
 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
1U12
 
 | | M. loti cyclic nucleotide binding domain mutant | | Descriptor: | IODIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-07-14 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Ligand Activation in a Cyclic Nucleotide Regulated Potassium Channel
Cell(Cambridge,Mass.), 119, 2004
|
|
1RGS
 
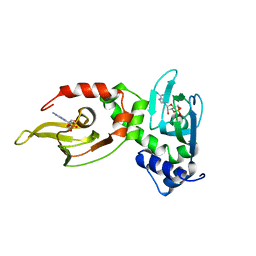 | | REGULATORY SUBUNIT OF CAMP DEPENDENT PROTEIN KINASE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP DEPENDENT PROTEIN KINASE | | Authors: | Su, Y, Dostmann, W.R.G, Herberg, F.W, Durick, K, Xuong, N.-H, Ten Eyck, L, Taylor, S.S, Varughese, K.I. | | Deposit date: | 1995-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains.
Science, 269, 1995
|
|
6CPB
 
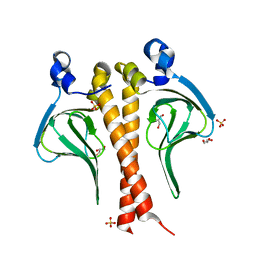 | |
6BQ8
 
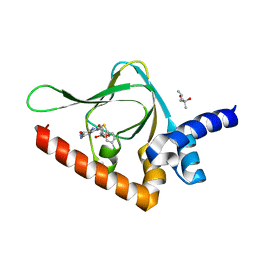 | | Joint X-ray/neutron structure of PKG II CNB-B domain in complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(~2~H_2_)amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2-hydroxy-7-(~2~H)hydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl](~2~H)-1,9-dihydro-6H-purin-6-one, STRONTIUM ION, ... | | Authors: | Kim, C, Kovalevsky, A, Gerlits, O. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallography Detects Differences in Protein Dynamics: Structure of the PKG II Cyclic Nucleotide Binding Domain in Complex with an Activator.
Biochemistry, 57, 2018
|
|
2K0G
 
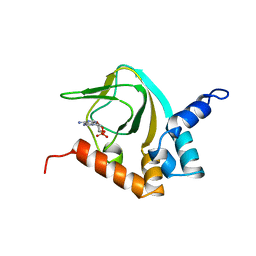 | |
7SSB
 
 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
4F8A
 
 | | Cyclic nucleotide binding-homology domain from mouse EAG1 potassium channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 1 | | Authors: | Marques-Carvalho, M.J, Sahoo, N, Muskett, F.W, Vieira-Pires, R.S, Gabant, G, Cadene, M, Schonherr, R, Morais-Cabral, J.H. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Biochemical, and Functional Characterization of the Cyclic Nucleotide Binding Homology Domain from the Mouse EAG1 Potassium Channel.
J.Mol.Biol., 423, 2012
|
|
3MDP
 
 | |
5V30
 
 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
