1K68
 
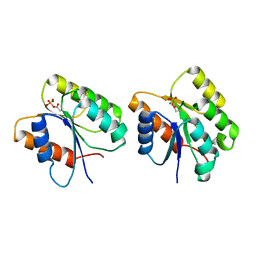 | | Crystal Structure of the Phosphorylated Cyanobacterial Phytochrome Response Regulator RcpA | | Descriptor: | MAGNESIUM ION, Phytochrome Response Regulator RcpA | | Authors: | Benda, C, Scheufler, C, Tandeau de Marsac, N, Gaertner, W. | | Deposit date: | 2001-10-15 | | Release date: | 2003-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of two cyanobacterial response regulators in apo- and phosphorylated form reveal a novel dimerization motif of phytochrome-associated response regulators
Biophys.J., 87, 2004
|
|
1K77
 
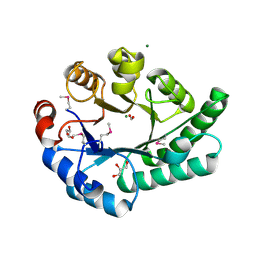 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1K90
 
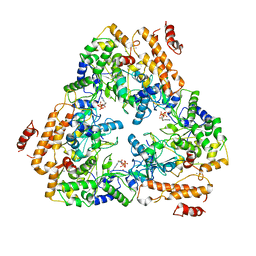 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
2AQS
 
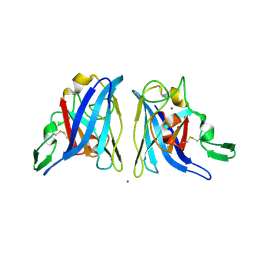 | | CU/ZN superoxide dismutase from neisseria meningitidis K91E, K94E double mutant | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Kassmann, C.J, Bruns, C.K, Cabelli, D.E, Cao, Z, Tabatabai, L.B, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 2005-08-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CU/ZN superoxide dismutase from neisseria meningitidis K91E, K94E double mutant
To be Published
|
|
1K3T
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi Complexed with Chalepin, a Coumarin Derivative Inhibitor | | Descriptor: | 6-(1,1-DIMETHYLALLYL)-2-(1-HYDROXY-1-METHYLETHYL)-2,3-DIHYDRO-7H-FURO[3,2-G]CHROMEN-7-ONE, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pavao, F. | | Deposit date: | 2001-10-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase complexed with chalepin, a natural product inhibitor, at 1.95 A resolution.
FEBS Lett., 520, 2002
|
|
2ARS
 
 | | Crystal structure of lipoate-protein ligase A From Thermoplasma acidophilum | | Descriptor: | Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ALV
 
 | | X-ray structural analysis of SARS coronavirus 3CL proteinase in complex with designed anti-viral inhibitors | | Descriptor: | N-((3S,6R)-6-((S,E)-4-ETHOXYCARBONYL-1-((S)-2-OXOPYRROLIDIN-3-YL)BUT-3-EN-2-YLCARBAMOYL)-2,9-DIMETHYL-4-OXODEC-8-EN-3-YL)-5-METHYLISOXAZOLE-3-CARBOXAMIDE, Replicase polyprotein 1ab | | Authors: | Ghosh, A.K, Xi, K, Ratia, K, Santarsiero, B.D, Fu, W, Harcourt, B.H, Rota, P.A, Baker, S.C, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2005-08-08 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors.
J.Med.Chem., 48, 2005
|
|
1K4G
 
 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
1K6E
 
 | | COMPLEX OF HYDROLYTIC HALOALKANE DEHALOGENASE LINB FROM SPHINGOMONAS PAUCIMOBILIS UT26 WITH 1,2-PROPANEDIOL (PRODUCT OF DEHALOGENATION OF 1,2-DIBROMOPROPANE) AT 1.85A | | Descriptor: | 1-BROMOPROPANE-2-OL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Streltsov, V.A, Prokop, Z, Damborsky, J, Nagata, Y, Oakley, A, Wilce, M.C.J. | | Deposit date: | 2001-10-16 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates.
Biochemistry, 42, 2003
|
|
1K6P
 
 | | LACK OF SYNERGY FOR INHIBITORS TARGETING A MULTI-DRUG RESISTANT HIV-1 PROTEASE | | Descriptor: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(BENZO[1,3]DIOXOL-5-YLMETHYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Schiffer, C.A. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease.
Protein Sci., 11, 2002
|
|
1K6W
 
 | | The Structure of Escherichia coli Cytosine Deaminase | | Descriptor: | Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
2B7M
 
 | |
2BB3
 
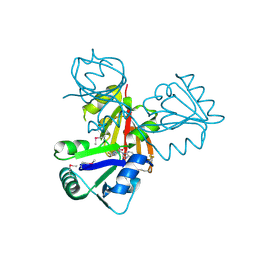 | | Crystal Structure of Cobalamin Biosynthesis Precorrin-6Y Methylase (cbiE) from Archaeoglobus fulgidus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, cobalamin biosynthesis precorrin-6Y methylase (cbiE) | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Cobalamin Biosynthesis Precorrin-6Y Methylase (cbiE) from Archaeoglobus fulgidus
To be Published
|
|
2ASI
 
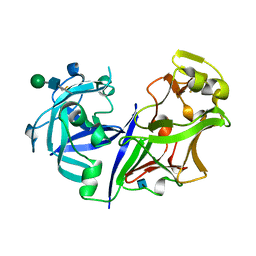 | | ASPARTIC PROTEINASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTIC PROTEINASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Jia, Z, Vandonselaar, M, Kepliakov, P.S.A, Quail, J.W. | | Deposit date: | 1995-12-09 | | Release date: | 1996-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 A resolution.
J.Mol.Biol., 268, 1997
|
|
1KAC
 
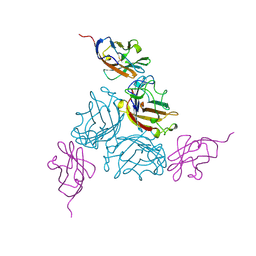 | | KNOB DOMAIN FROM ADENOVIRUS SEROTYPE 12 IN COMPLEX WITH DOMAIN 1 OF ITS CELLULAR RECEPTOR CAR | | Descriptor: | PROTEIN (COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR), PROTEIN (FIBER KNOB PROTEIN) | | Authors: | Bewley, M.C, Springer, K, Zhang, Y.B, Freimuth, P, Flanagan, J.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR.
Science, 286, 1999
|
|
1KA4
 
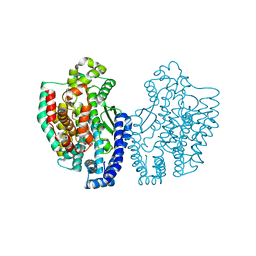 | | Structure of Pyrococcus furiosus carboxypeptidase Nat-Pb | | Descriptor: | LEAD (II) ION, M32 carboxypeptidase | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
1CWD
 
 | | HUMAN P56LCK TYROSINE KINASE COMPLEXED WITH PHOSPHOPEPTIDE | | Descriptor: | (PHOSPHONOMETHYL)PHENYLALANINE-CONTAINING PEPTIDE PRO-GLU-GLY-ASP-PM3-GLU-GLU-VAL-LEU, P56LCK TYROSINE KINASE | | Authors: | Mikol, V. | | Deposit date: | 1995-09-06 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structures of the SH2 domain of p56lck complexed with two phosphopeptides suggest a gated peptide binding site.
J.Mol.Biol., 246, 1995
|
|
2AJ9
 
 | | X-ray crystal structure of W42A,R161A double mutant of Mycobacterium tuberculosis beta-ketoacyl-ACP synthase III | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III | | Authors: | Brown, A.K, Sridharan, S, Kremer, L, Lindenberg, S, Dover, L.G, Sacchettini, J.C, Besra, G.S. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of the Mycobacterium tuberculosis beta-Ketoacyl-Acyl Carrier Protein Synthase III mtFabH: Factors Influencing Catalysis And Substrate Specificity.
J.Biol.Chem., 280, 2005
|
|
1K66
 
 | | Crystal Structure of the Cyanobacterial Phytochrome Response Regulator, RcpB | | Descriptor: | BETA-MERCAPTOETHANOL, Phytochrome Response Regulator RcpB | | Authors: | Benda, C, Scheufler, C, Tandeau de Marsac, N, Gaertner, W. | | Deposit date: | 2001-10-15 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of two cyanobacterial response regulators in apo- and phosphorylated form reveal a novel dimerization motif of phytochrome-associated response regulators
Biophys.J., 87, 2004
|
|
2AY8
 
 | |
1CWE
 
 | | HUMAN P56LCK TYROSINE KINASE COMPLEXED WITH PHOSPHOPEPTIDE | | Descriptor: | P56LCK TYROSINE KINASE, PHOSPHOPEPTIDE ACQ-PMP-GLU-GLU-ILE-PRO | | Authors: | Mikol, V. | | Deposit date: | 1995-09-06 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of the SH2 domain of p56lck complexed with two phosphopeptides suggest a gated peptide binding site.
J.Mol.Biol., 246, 1995
|
|
2AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14Q | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-18 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1K61
 
 | | MATALPHA2 HOMEODOMAIN BOUND TO DNA | | Descriptor: | 5'-D(*(5IU)P*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*AP*CP*AP*TP*G)-3', 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*CP*AP*CP*GP*C)-3', Mating-type protein alpha-2 | | Authors: | Aishima, J, Gitti, R.K, Noah, J.E, Gan, H.H, Schlick, T, Wolberger, C. | | Deposit date: | 2001-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Hoogsteen base pair embedded in undistorted B-DNA
NUCLEIC ACIDS RES., 30, 2002
|
|
2ANM
 
 | | Ternary complex of an orally active thrombin inhibitor with human thrombin and a c-terminal hirudin derived exo-sit inhibitor | | Descriptor: | 2-((R)-1-((S)-2-(N-(6-CARBAMIMIDOYLPYRIDIN-3-YL)METHYLCARBAMOYL)-2H-PYRROL-1(5H)-YL)-3-CYCLOHEXYL-1-OXOPROPAN-2-YLAMINO)ACETIC ACID, thrombin | | Authors: | Lange, U.E.W, Baucke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-06-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Orally active thrombin inhibitors. Part 2: optimization of the P2-moiety
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2ANZ
 
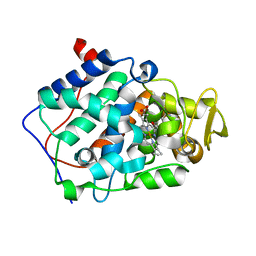 | | cytochrome c peroxidase in complex with 2,6-diaminopyridine | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-08-11 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
