1UCF
 
 | | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Honbou, K, Suzuki, N.N, Horiuchi, M, Niki, T, Taira, T, Ariga, H, Inagaki, F. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease
J.BIOL.CHEM., 278, 2003
|
|
1BGV
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
3M7S
 
 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
9IZU
 
 | | Cryo-EM structure of ALDH6A1-P62S | | Descriptor: | Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase [acylating], mitochondrial | | Authors: | Su, G, Xu, Y, Luan, X. | | Deposit date: | 2024-08-01 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and biochemical basis for the pathogenic mutations of methylmalonate semialdehyde dehydrogenase ALDH6A1
Medicine Plus, 1, 2024
|
|
1UBE
 
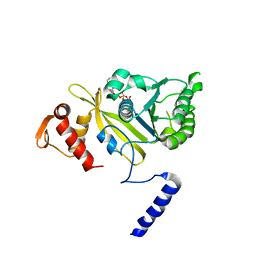 | | MsRecA-ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
8P1Q
 
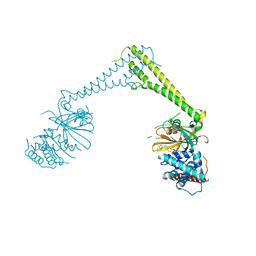 | | USP28 in complex with FT206 | | Descriptor: | 3-azanyl-N-[(2S)-6-[(1S,5R)-3,8-diazabicyclo[3.2.1]octan-3-yl]-1,2,3,4-tetrahydronaphthalen-2-yl]-6-methyl-thieno[2,3-b]pyridine-2-carboxamide, DIMETHYL SULFOXIDE, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Sauer, F, Karal Nair, R, Kisker, C. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for the bi-specificity of USP25 and USP28 inhibitors.
Embo Rep., 25, 2024
|
|
1UD8
 
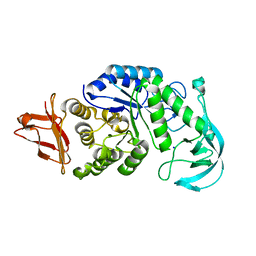 | | Crystal structure of AmyK38 with lithium ion | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD3
 
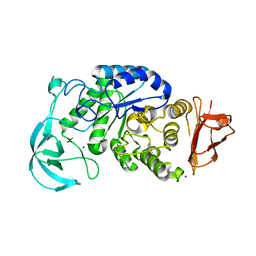 | | Crystal structure of AmyK38 N289H mutant | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
5H73
 
 | | Crystal structure of human DHODH with 18F | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Huang, J, Wu, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of human DHODH with 18F at 1.58 Angstroms resolution
To Be Published
|
|
1UDY
 
 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
1UG6
 
 | | Structure of beta-glucosidase at atomic resolution from thermus thermophilus HB8 | | Descriptor: | GLYCEROL, beta-glycosidase | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-12 | | Release date: | 2003-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of Beta-Glucosidase at Atomic Resolution from Thermus Thermophilus Hb8
To be Published
|
|
3MDT
 
 | | Voriconazole complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
1AXB
 
 | |
5H8W
 
 | | XPD mechanism | | Descriptor: | ATP-dependent DNA helicase Ta0057, DNA (5'-D(P*TP*AP*CP*GP*A)-3'), IRON/SULFUR CLUSTER, ... | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
3MGW
 
 | | Thermodynamics and structure of a salmon cold-active goose-type lysozyme | | Descriptor: | COBALT (II) ION, Lysozyme g, SULFATE ION | | Authors: | Kyomuhendo, P, Myrnes, B, Brandsdal, B.O, Smalas, A.O, Nilsen, I.W, Helland, R. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics and structure of a salmon cold active goose-type lysozyme
Comp.Biochem.Physiol. B: Biochem.Mol.Biol., 156, 2010
|
|
1BKW
 
 | | p-Hydroxybenzoate hydroxylase (phbh) mutant with cys116 replaced by ser (c116s) and arg44 replaced by lys (r44k), in complex with fad and 4-hydroxybenzoic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H, Schreuder, H.A, Van Berkel, W.J. | | Deposit date: | 1998-07-13 | | Release date: | 1998-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
Eur.J.Biochem., 231, 1995
|
|
3MI6
 
 | | Crystal structure of the alpha-galactosidase from Lactobacillus brevis, Northeast Structural Genomics Consortium Target LbR11. | | Descriptor: | Alpha-galactosidase | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Belote, R, Sahdev, S, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of the alpha-galactosidase from Lactobacillus brevis.
To be Published
|
|
3MJL
 
 | |
3MJY
 
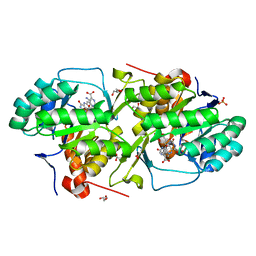 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Aminoorotic acid | | Descriptor: | 5-amino-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-13 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
3MK7
 
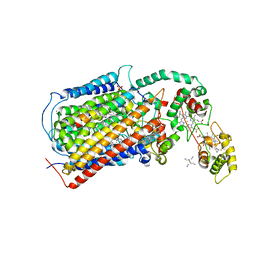 | | The structure of CBB3 cytochrome oxidase | | Descriptor: | 30-mer peptide, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Buschmann, S, Warkentin, E, Michel, H, Ermler, U. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of cbb3 Cytochrome Oxidase Provides Insights into Proton Pumping
Science, 329, 2010
|
|
5HDI
 
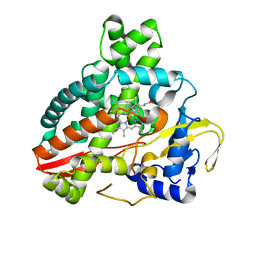 | | Structural characterization of CYP144A1, a Mycobacterium tuberculosis cytochrome P450 | | Descriptor: | Cytochrome P450 144, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chenge, J, Driscoll, M.D, McLean, K.J, Munro, A.W, Leys, D. | | Deposit date: | 2016-01-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural characterization of CYP144A1 - a cytochrome P450 enzyme expressed from alternative transcripts in Mycobacterium tuberculosis.
Sci Rep, 6, 2016
|
|
3MHU
 
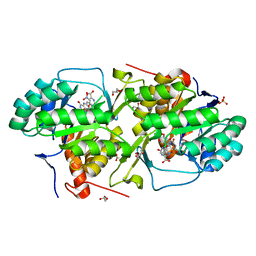 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Nitroorotic acid | | Descriptor: | 5-nitro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
5HDR
 
 | | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine | | Descriptor: | Alpha-L-fucosidase, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2016-01-05 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine
To Be Published
|
|
8K1A
 
 | | the wild-typed alpha-galactosidase 5 | | Descriptor: | Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activity-based metaproteomics driven discovery and enzymological characterization of potential alpha-galactosidases in the mouse gut microbiome.
Commun Chem, 7, 2024
|
|
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
