2KXP
 
 | |
2GTJ
 
 | |
2GTO
 
 | |
2K1S
 
 | | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553. | | Descriptor: | Inner membrane lipoprotein yiaD | | Authors: | Ramelot, T.A, Zhao, L, Hamilton, K, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553.
To be Published
|
|
2FBS
 
 | |
1ZG2
 
 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
2ERM
 
 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
2JVD
 
 | | Solution NMR structure of the folded N-terminal fragment of UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384-1-46 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Sharma, S, Huang, Y.J, Zhao, L, Owens, L.A, Stokes, K, Jiang, M, Xiao, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis.
Proteins, 72, 2008
|
|
1E9K
 
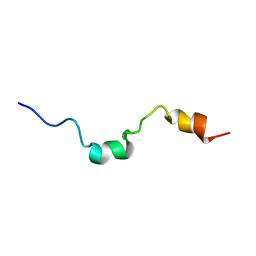 | | The structure of the RACK1 interaction sites located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. | | Descriptor: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Bolger, G.B, Smith, K.J, McCahill, A, Hyde, E.I, Steele, M.R, Houslay, M.D. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1.
Cell. Signal., 19, 2007
|
|
2HEP
 
 | | Solution NMR structure of the UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384. | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Swapna, G.V.T, Ho, C.K, Shetty, K, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis
Proteins, 72, 2008
|
|
2JRS
 
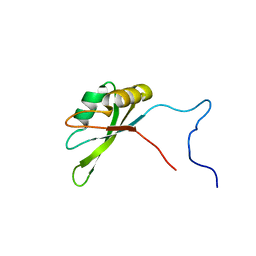 | | Solution NMR Structure of CAPER RRM2 Domain. Northeast Structural Genomics Target HR4730A | | Descriptor: | RNA-binding protein 39 | | Authors: | Rossi, P, Zhao, L, Nwosu, C, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CAPER RRM2 Domain.
To be Published
|
|
1SR3
 
 | |
2MLE
 
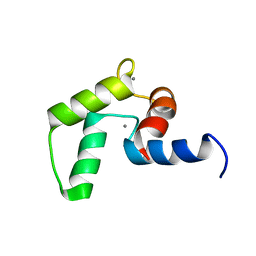 | | NMR structure of the C-domain of troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Baryshnikova, O.K, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
2MLF
 
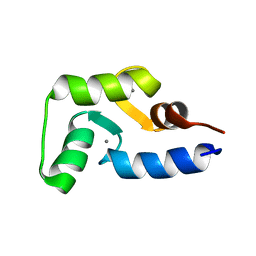 | | NMR structure of the dilated cardiomyopathy mutation G159D in troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Baryshnikova, O.K, Robertson, I.M, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
1T17
 
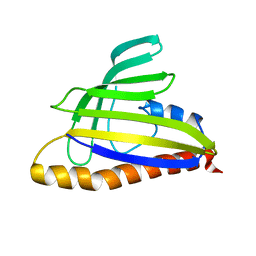 | | Solution Structure of the 18 kDa Protein CC1736 from Caulobacter crescentus: The Northeast Structural Genomics Consortium Target CcR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Shen, Y, Atreya, H.S, Acton, T, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity.
Proteins, 58, 2005
|
|
2JQF
 
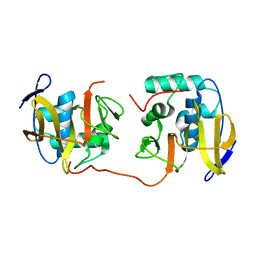 | | Full Length Leader Protease of Foot and Mouth Disease Virus C51A Mutant | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
1YWL
 
 | |
2JQG
 
 | | Leader Protease | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
2KJN
 
 | |
2KJO
 
 | |
1LES
 
 | | LENTIL LECTIN COMPLEXED WITH SUCROSE | | Descriptor: | CALCIUM ION, LENTIL LECTIN, MANGANESE (II) ION, ... | | Authors: | Hamelryck, T, Loris, R. | | Deposit date: | 1995-08-23 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NMR, molecular modeling, and crystallographic studies of lentil lectin-sucrose interaction.
J.Biol.Chem., 270, 1995
|
|
2HFI
 
 | | Solution NMR Structure of Protein yppE from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR213 | | Descriptor: | Hypothetical protein yppE | | Authors: | Liu, G, Singarapu, K.K, Parish, D, Eletsky, A, Xu, D, Sukumaran, D, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of hypothetical protein yppE: Northeast Structural Genomics Consortium Target SR213
TO BE PUBLISHED
|
|
2G7J
 
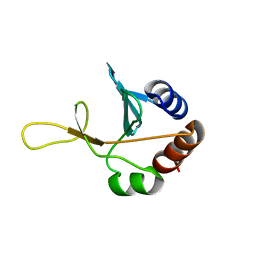 | | Solution NMR structure of the putative cytoplasmic protein ygaC from Salmonella typhimurium. Northeast Structural Genomics target StR72. | | Descriptor: | putative cytoplasmic protein | | Authors: | Aramini, J.M, Swapna, G.V.T, Ramelot, T.A, Ho, C.K, Cunningham, K, Ma, L.-C, Shetty, K, Xiao, R, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the putative cytoplasmic protein ygaC from Salmonella typhimurium. Northeast Structural Genomics target StR72.
To be Published
|
|
2JS7
 
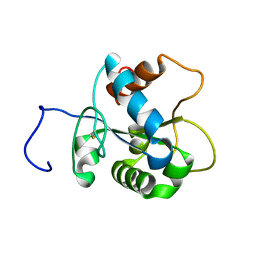 | | Solution NMR structure of human myeloid differentiation primary response (MyD88). Northeast Structural Genomics target HR2869A | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Rossi, P, Ramelot, T.A, Tao, X, Ciano, M, Ho, C, Ma, L.-C, Xiao, R, Acton, T.B, Kennedy, M.A, Tong, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Myeloid Differentiation Primary Response (MyD88).
To be Published
|
|
2JU6
 
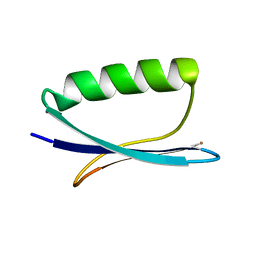 | | Solid-State Protein Structure Determination with Proton-Detected Triple Resonance 3D Magic-Angle Spinning NMR Spectroscopy | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Zhou, D.H, Shea, J.J, Nieuwkoop, A.J, Franks, W, Wylie, B.J, Mullen, C, Sandoz, D, Rienstra, C.M. | | Deposit date: | 2007-08-15 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | Solid-State Protein-Structure Determination with Proton-Detected Triple-Resonance 3D Magic-Angle-Spinning NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
