4Y2U
 
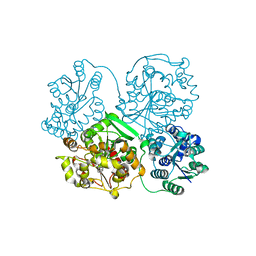 | | Structure of soluble epoxide hydrolase in complex with tert-butyl 1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, tert-butyl (3R)-1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
6ABH
 
 | |
4XKW
 
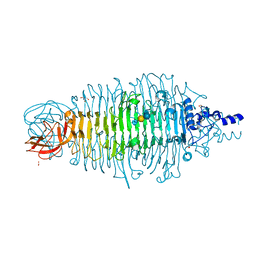 | | Tailspike protein mutant D339N of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-12 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XLA
 
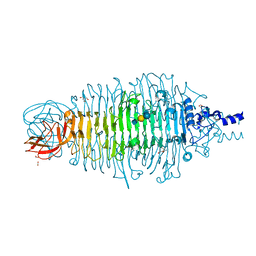 | | Tailspike protein mutant D339A of E. coli bacteriophage HK620 IN COMPLEX WITH PENTASACCHARIDE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XM3
 
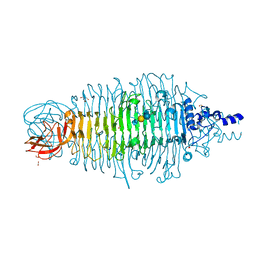 | | Tailspike protein mutant E372A of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XN3
 
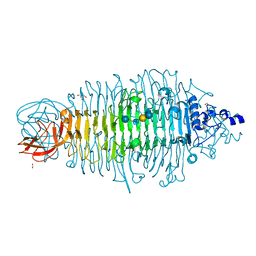 | | Tailspike protein mutant E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
2MUR
 
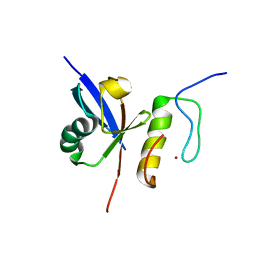 | |
2MUB
 
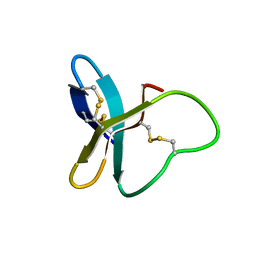 | |
2N19
 
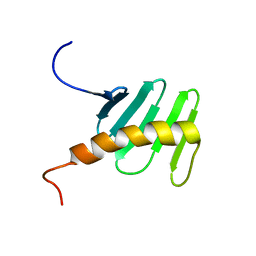 | | STIL binding to the Polo-box domain 3 of PLK4 regulates centriole duplication | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Boehm, R, Arquint, C, Gabryjonczyk, A, Imseng, S, Sauer, E, Hiller, S, Nigg, E, Maier, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | STIL binding to Polo-box 3 of PLK4 regulates centriole duplication.
Elife, 4, 2015
|
|
4XON
 
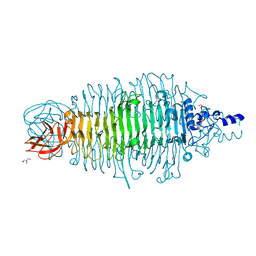 | | Tailspike protein double mutant D339N/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
2N3D
 
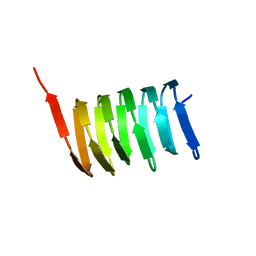 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
2N1F
 
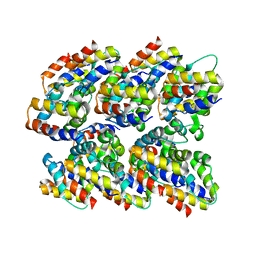 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6AB8
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZLys | | Descriptor: | (2S)-2-azanyl-6-(phenylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
2N7I
 
 | |
4XQB
 
 | | CRYSTAL STRUCTURE OF AD37 FIBER KNOB IN COMPLEX WITH TRIVALENT SIALIC ACID INHIBITOR ME0461 | | Descriptor: | 2-(1-{2-[bis(2-{4-[2-({(6R)-5-(acetylamino)-3,5-dideoxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-beta-L-threo-hex-2-ulopyranonosyl}oxy)ethyl]-1H-1,2,3-triazol-1-yl}ethyl)amino]ethyl}-1H-1,2,3-triazol-4-yl)ethyl (6R)-5-(acetylamino)-3,5-dideoxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-beta-L-threo-hex-2-ulopyranosidonic acid, ACETATE ION, Fiber, ... | | Authors: | Stehle, T, Liaci, A.M. | | Deposit date: | 2015-01-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
4XWY
 
 | | Crystal structure of human sepiapterin reductase in complex with an N-acetylserotinin analogue | | Descriptor: | N-[2-(5-hydroxy-2-methyl-1H-indol-3-yl)ethyl]-2-methoxyacetamide, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Johnsson, K, Hovius, R, Gorszka, K.I, Pojer, F. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Reduction of Neuropathic and Inflammatory Pain through Inhibition of the Tetrahydrobiopterin Pathway.
Neuron, 86, 2015
|
|
2N7M
 
 | | NMR-SAXS/WAXS Structure of the core of the U4/U6 di-snRNA | | Descriptor: | RNA (92-MER) | | Authors: | Cornilescu, G, Didychuk, A.L, Rodgers, M.L, Michael, L.A, Burke, J.E, Montemayor, E.J, Hoskins, A.A, Butcher, S.E, Tonelli, M. | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA.
J.Mol.Biol., 428, 2016
|
|
4XR6
 
 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
6AEI
 
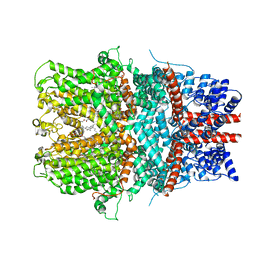 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-08-05 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
5U27
 
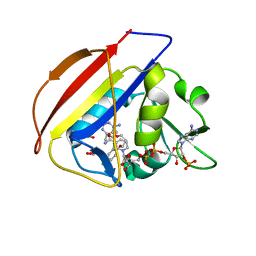 | |
2MV2
 
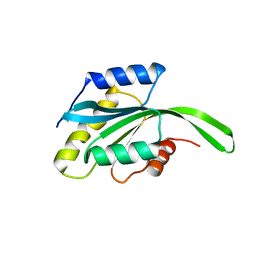 | |
2N3K
 
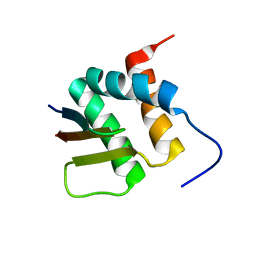 | | Human Brd4 ET domain in complex with MLV Integrase C-term | | Descriptor: | Bromodomain-containing protein 4, MLV integrase | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5U5U
 
 | | CcP gateless cavity | | Descriptor: | ISONICOTINAMIDINE, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2MZ8
 
 | | Solution NMR structure of Salmonella Typhimurium transcriptional regulator protein Crl | | Descriptor: | Sigma factor-binding protein Crl | | Authors: | Cavaliere, P, Levi-Acobas, F, Monteil, V, Bellalou, J, Mayer, C, Norel, F, Sizun, C. | | Deposit date: | 2015-02-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding interface between the Salmonella sigma (S)/RpoS subunit of RNA polymerase and Crl: hints from bacterial species lacking crl.
Sci Rep, 5, 2015
|
|
5U5Y
 
 | | CcP gateless cavity | | Descriptor: | 2-methylimidazo[1,2-a]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
