6FI7
 
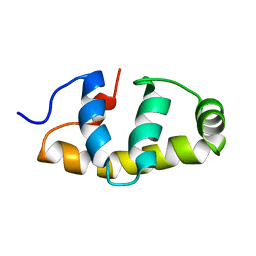 | | E.coli Sigma factor S (RpoS) Region 4 | | Descriptor: | RNA polymerase sigma factor RpoS | | Authors: | Liu, B, Matthews, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | T7 phage factor required for managing RpoS inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8FZJ
 
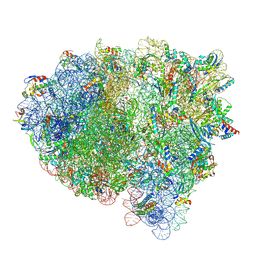 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZG
 
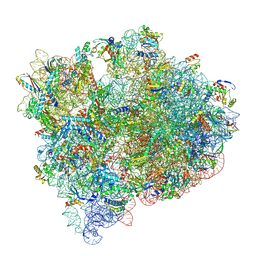 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZF
 
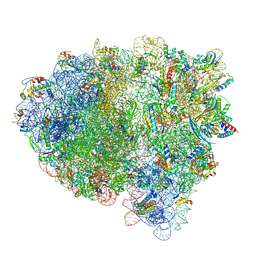 | | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZE
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZH
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8G7R
 
 | |
8G7S
 
 | |
5HDG
 
 | |
1HOM
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Qian, Y.-Q, Billeter, M, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
5SYT
 
 | | Crystal Structure of ZMPSTE24 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX prenyl protease 1 homolog, ... | | Authors: | Clark, K, Jenkins, J.L, Fedoriw, N, Dumont, M.E, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Protein Sci., 26, 2017
|
|
2CHQ
 
 | | Replication Factor C ADPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
5K4J
 
 | | Crystal Structure of CDK2 in complex with compound 22 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Cyclin-dependent kinase 2 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
8P2K
 
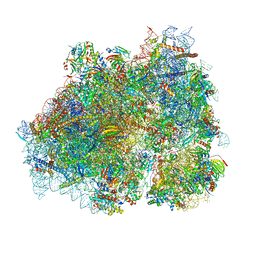 | | Ternary complex of translating ribosome, NAC and METAP1 | | Descriptor: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | Deposit date: | 2023-05-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
4GIZ
 
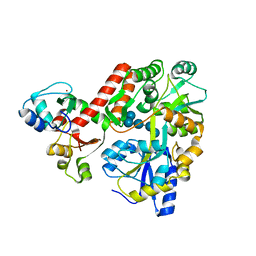 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
5JQ5
 
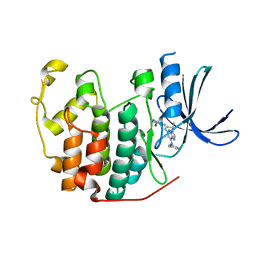 | | Crystal structure of CDK2 in complex with inhibitor ICEC0942 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, ACETATE ION, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
5JQ8
 
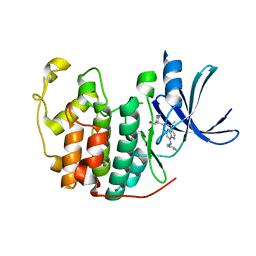 | | Crystal structure of CDK2 in complex with inhibitor ICEC0943 | | Descriptor: | (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
6ZRD
 
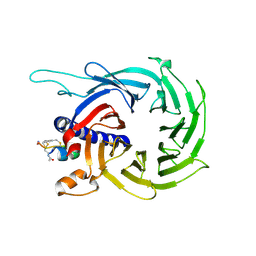 | |
5IEV
 
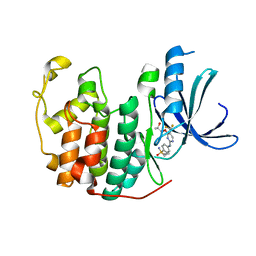 | | Crystal structure of BAY 1000394 (Roniciclib) bound to CDK2 | | Descriptor: | Cyclin-dependent kinase 2, Roniciclib | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5L2W
 
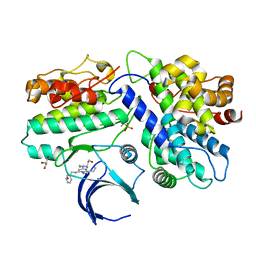 | | The X-ray co-crystal structure of human CDK2/CyclinE and Dinaciclib. | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1, ... | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
2KNH
 
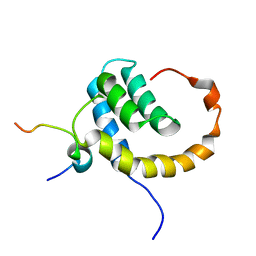 | | The Solution structure of the eTAFH domain of AML1-ETO complexed with HEB peptide | | Descriptor: | Protein CBFA2T1, Transcription factor 12 | | Authors: | Park, S, Cierpicki, T, Tonelli, M, Bushweller, J.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity.
Blood, 113, 2009
|
|
3SDG
 
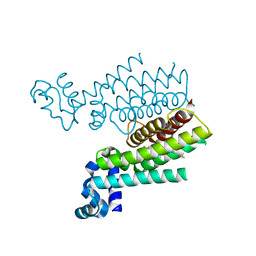 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 4,4,4-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}butan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
2F8D
 
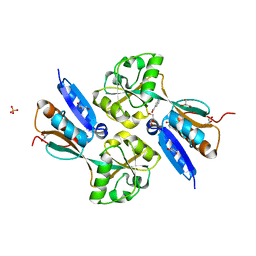 | | BenM effector-Binding domain crystallized from high pH conditions | | Descriptor: | BENZOIC ACID, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ezezika, O.C, Haddad, S, Neidle, E.L, Momany, C. | | Deposit date: | 2005-12-02 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oligomerization of BenM, a LysR-type transcriptional regulator: structural basis for the aggregation of proteins in this family.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2F4U
 
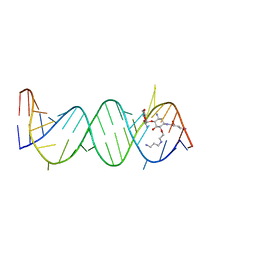 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-((1R,2S,3R,4R,5S)-5-AMINO-4-[(2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-{2-[(3-AMINOPROPYL)AMINO]ETHOXY}-3-HYDROXYCYCLOHEXYL)-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2FD0
 
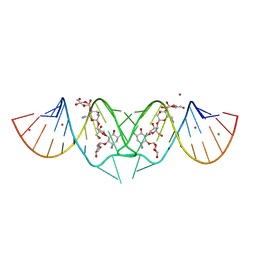 | | HIV-1 DIS kissing-loop in complex with lividomycin | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
