1C41
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, LUMAZINE SYNTHASE, SULFATE ION | | Authors: | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly
Protein Sci., 8, 1999
|
|
1C83
 
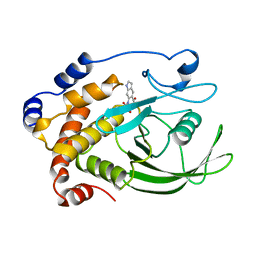 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 6-(OXALYL-AMINO)-1H-INDOLE-5-CARBOXYLIC ACID | | Descriptor: | 6-(OXALYL-AMINO)-1H-INDOLE-5-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P. | | Deposit date: | 2000-04-14 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1C1H
 
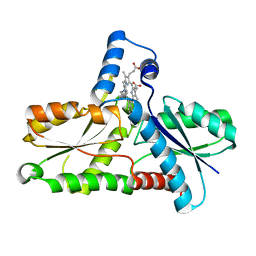 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS FERROCHELATASE IN COMPLEX WITH N-METHYL MESOPORPHYRIN | | Descriptor: | FERROCHELATASE, MAGNESIUM ION, N-METHYLMESOPORPHYRIN | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1BXE
 
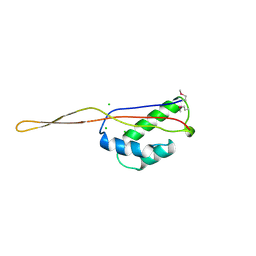 | | RIBOSOMAL PROTEIN L22 FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, PROTEIN (RIBOSOMAL PROTEIN L22) | | Authors: | Unge, J, Aberg, A, Al-Karadaghi, S, Nikulin, A, Nikonov, S, Davydova, N, Nevskaya, N, Garber, M, Liljas, A. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance.
Structure, 6, 1998
|
|
1A8Z
 
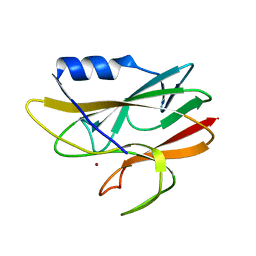 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1CJS
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 50S RIBOSOMAL PROTEIN L1P | | Authors: | Nevskaya, N, Tishchenko, S, Fedorov, R, Al-Karadaghi, S, Liljas, A, Kraft, A, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 1999-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family.
Structure Fold.Des., 8, 2000
|
|
1CV2
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 AT 1.6 A resolution | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-08-22 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
1CQM
 
 | |
1C86
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B (R47V,D48N) COMPLEXED WITH 2-(OXALYL-AMINO-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1DD5
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA RIBOSOME RECYCLING FACTOR, RRF | | Descriptor: | ACETIC ACID, RIBOSOME RECYCLING FACTOR | | Authors: | Selmer, M, Al-Karadaghi, S, Hirokawa, G, Kaji, A, Liljas, A. | | Deposit date: | 1999-11-08 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic.
Science, 286, 1999
|
|
1B2L
 
 | | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS: TERNARY COMPLEX WITH NAD-CYCLOHEXANONE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ALCOHOL DEHYDROGENASE, CALCIUM ION, ... | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1B3N
 
 | | BETA-KETOACYL CARRIER PROTEIN SYNTHASE AS A DRUG TARGET, IMPLICATIONS FROM THE CRYSTAL STRUCTURE OF A COMPLEX WITH THE INHIBITOR CERULENIN. | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, PROTEIN (KETOACYL ACYL CARRIER PROTEIN SYNTHASE 2) | | Authors: | Moche, M, Schneider, G, Edwards, P, Dehesh, K, Lindqvist, Y. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase.
J.Biol.Chem., 274, 1999
|
|
1DOZ
 
 | | CRYSTAL STRUCTURE OF FERROCHELATASE | | Descriptor: | FERROCHELATASE, MAGNESIUM ION | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-12-22 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1D07
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
1E3L
 
 | | P47H mutant of mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | ALCOHOL DEHYDROGENASE, CLASS II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-19 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3E
 
 | | Mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoeoeg, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-14 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3I
 
 | | Mouse class II alcohol dehydrogenase complex with NADH and inhibitor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-16 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1ENF
 
 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 1.69 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1EO9
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE AT PH < 7.0 | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, PROTOCATECHUATE 3,4-DIOXYGENASE BETA CHAIN;, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EOB
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
3LSK
 
 | | Pyranose 2-oxidase T169S acetate complex | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-25 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | H-bonding and positive charge at the N5/O4 locus are critical for covalent flavin attachment in trametes pyranose 2-oxidase.
J.Mol.Biol., 402, 2010
|
|
3LSH
 
 | | Pyranose 2-oxidase T169A, monoclinic | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Divne, C, Tan, T.C, Spadiut, O. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H-bonding and positive charge at the N5/O4 locus are critical for covalent flavin attachment in trametes pyranose 2-oxidase.
J.Mol.Biol., 402, 2010
|
|
3M4Z
 
 | | Crystal Structure of B. subtilis ferrochelatase with Cobalt bound at the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferrochelatase, ... | | Authors: | Soderberg, C.A.G, Hansson, M.D, Sreekanth, R, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Bacterial ferrochelatase goes human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase
To be Published
|
|
6VFD
 
 | | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site.
To be Published
|
|
6NV6
 
 | |
