6QLO
 
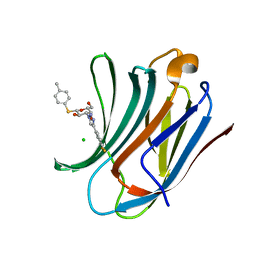 | | Galectin-3C in complex with substituted fluoroaryltriazole monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[3,4-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6QLQ
 
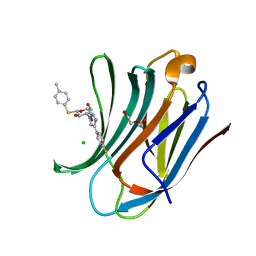 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative 4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6QLR
 
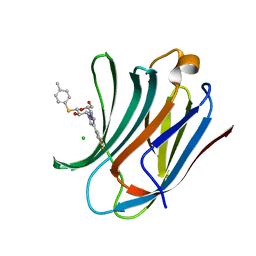 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative 5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6QLN
 
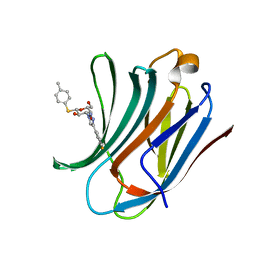 | | Galectin-3C in complex with fluoroaryl triazole monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6QLU
 
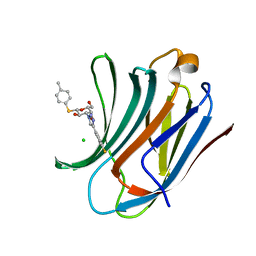 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(4-fluoranyl-3-methyl-phenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6R2W
 
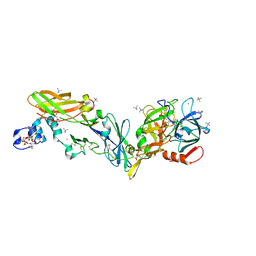 | | Crystal structure of the super-active FVIIa variant VYT in complex with tissue factor | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-acetyl-D-phenylalanyl-N-[(2S,3S)-6-carbamimidamido-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Beating tissue factor at its own game: Design and properties of a soluble tissue factor-independent coagulation factor VIIa.
J.Biol.Chem., 295, 2020
|
|
6R71
 
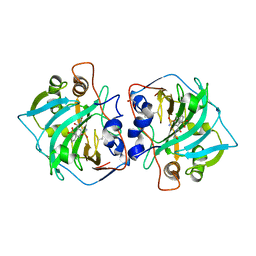 | | Crystal structure of human carbonic anhydrase isozyme XII with 2-(benzenesulfonyl)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-(phenylsulfonyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
6R6J
 
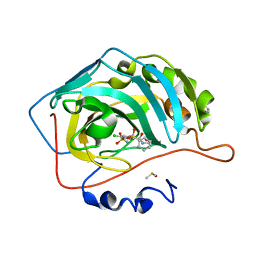 | | Crystal structure of human carbonic anhydrase isozyme II with 2-(benzenesulfonyl)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-(phenylsulfonyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
6RS6
 
 | | X-ray crystal structure of LsAA9B | | Descriptor: | AA9, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
2G4F
 
 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | Hydrolase | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2GD0
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (S)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GBT
 
 | | C6A/C111A CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2GFT
 
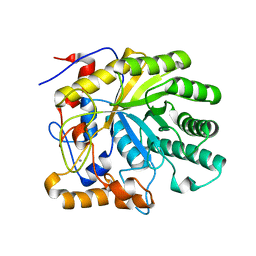 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
2GBU
 
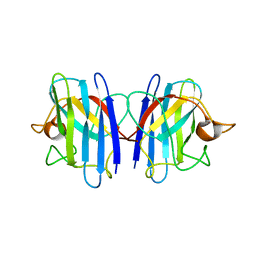 | | C6A/C111A/C57A/C146A apo CuZn Superoxide dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2GBV
 
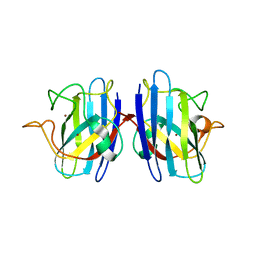 | | C6A/C111A/C57A/C146A holo CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2GCI
 
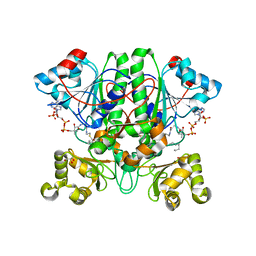 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an asparte/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GIM
 
 | |
6Z2T
 
 | |
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1C84
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 3-(OXALYL-AMINO)-NAPHTHALENE-2-CARBOXLIC ACID | | Descriptor: | 3-(OXALYL-AMINO)-NAPHTHALENE-2-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P. | | Deposit date: | 2000-04-14 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1C2Y
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PROTEIN (LUMAZINE SYNTHASE) | | Authors: | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | Deposit date: | 1999-07-27 | | Release date: | 2000-07-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly.
Protein Sci., 8, 1999
|
|
1C87
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1C85
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO)-BENZOIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-BENZOIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1C88
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
