8UNM
 
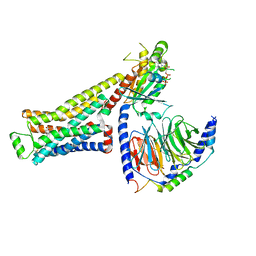 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class B) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNW
 
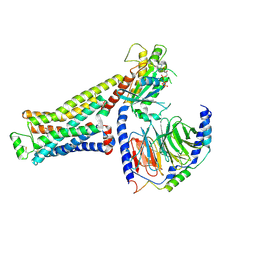 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class L) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
3JUX
 
 | |
6YW8
 
 | |
6HTS
 
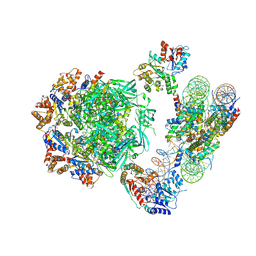 | | Cryo-EM structure of the human INO80 complex bound to nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Ayala, R, Willhoft, O, Aramayo, R.J, Wilkinson, M, McCormack, E.A, Ocloo, L, Wigley, D.B, Zhang, X. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and regulation of the human INO80-nucleosome complex.
Nature, 556, 2018
|
|
3JV2
 
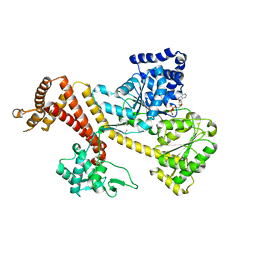 | | Crystal Structure of B. subtilis SecA with bound peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translocase subunit SecA, ... | | Authors: | Zimmer, J. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility and peptide interaction of the translocation ATPase SecA.
J.Mol.Biol., 394, 2009
|
|
6K72
 
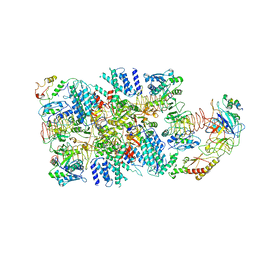 | | eIF2(aP) - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6K71
 
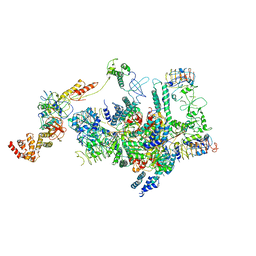 | | eIF2 - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
7Y7L
 
 | | Solution structure of zinc finger domain 2 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-22 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7YAB
 
 | | Solution structure of zinc finger domain 1 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
4EIG
 
 | | CA1698 camel antibody fragment in complex with DHFR | | Descriptor: | CA1698 camel antibody fragment, Dihydrofolate reductase | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
4EIZ
 
 | | Structure of Nb113 bound to apoDHFR | | Descriptor: | Dihydrofolate reductase, Nb113 Camel antibody fragment | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-06 | | Release date: | 2013-04-24 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
8DZH
 
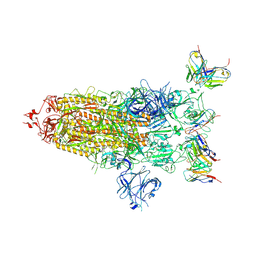 | |
5X0Y
 
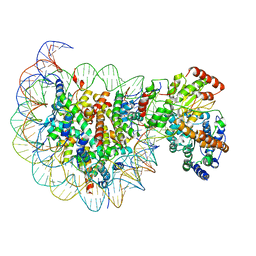 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
5X26
 
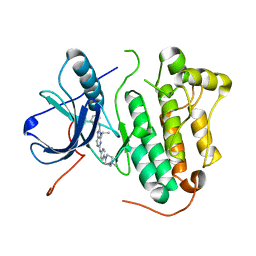 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
4QHN
 
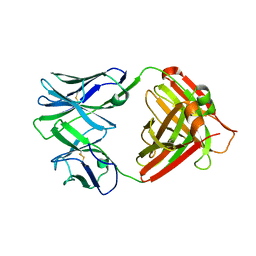 | | I2 (unbound) from CH103 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5X2K
 
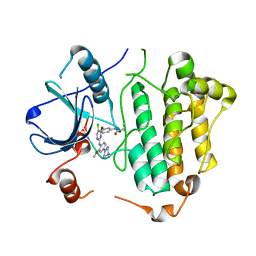 | | Crystal structure of EGFR 696-1022 T790M in complex with WZ4003 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Zhu, S.J, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5W88
 
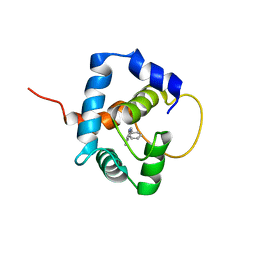 | |
5WCL
 
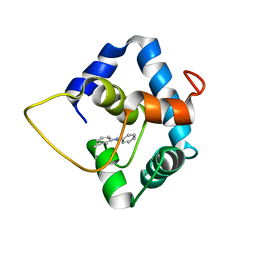 | | NMR structure of the N-domain of troponin C bound to switch region of troponin I and 3-methyldiphenylamine (solvent exposed mode) | | Descriptor: | 3-methyl-N-phenylaniline, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures reveal details of small molecule binding to cardiac troponin.
J. Mol. Cell. Cardiol., 101, 2016
|
|
3TSD
 
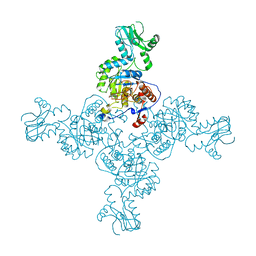 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
4QI9
 
 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
5WCV
 
 | |
3TSB
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
