7BKP
 
 | | CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Singh, R, Herrero del Valle, A, Yu, Q, Modis, Y. | | Deposit date: | 2021-01-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
7BAI
 
 | | Structure of RIG-I CTD (I875A) bound to p-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*(GDP)P*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved isoleucine in the binding pocket of RIG-I controls immune tolerance to mitochondrial RNA.
Nucleic Acids Res., 51, 2023
|
|
7BAH
 
 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the binding pocket of RIG-I controls immune tolerance to mitochondrial RNA.
Nucleic Acids Res., 51, 2023
|
|
2F4T
 
 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
7Y80
 
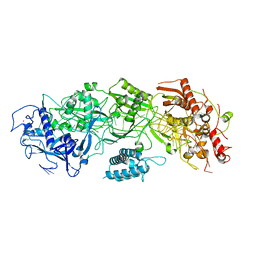 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA binary complex | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, ZINC ION, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y84
 
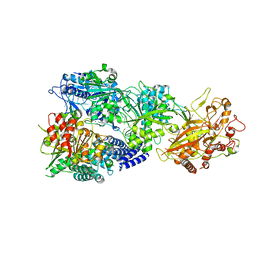 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
2KJM
 
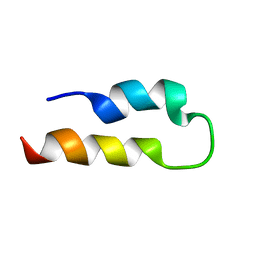 | | Solution structure of SLBP RNA binding domain fragment | | Descriptor: | Histone RNA hairpin-binding protein | | Authors: | Thapar, R. | | Deposit date: | 2009-06-03 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP-RNA complex by phosphorylation and proline isomerization.
Biochemistry, 51, 2012
|
|
6J7Y
 
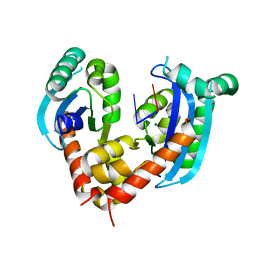 | | Human mitochondrial Oligoribonuclease in complex with DNA | | Descriptor: | DNA (5'-D(P*TP*T)-3'), MAGNESIUM ION, Oligoribonuclease, ... | | Authors: | Chu, L.Y, Agrawal, S, Yuan, H.S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Structural insights into nanoRNA degradation by human Rexo2.
Rna, 25, 2019
|
|
7SOW
 
 | | LaM domain of human LARP1 in complex with UUUUUU | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of La-related protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Enhanced binding of guanylated poly(A) RNA by the LaM domain of LARP1.
Rna Biol., 21, 2024
|
|
7BKQ
 
 | | CryoEM structure of MDA5-dsRNA filament in complex with ADP with 92-degree helical twist | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), ... | | Authors: | Yu, Q, Modis, Y. | | Deposit date: | 2021-01-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
3CDI
 
 | | Crystal structure of E. coli PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
7CBH
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | Threonine--tRNA ligase, ZINC ION, [(E)-4-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)but-2-enyl] (2S,3R)-2-azanyl-3-oxidanyl-butanoate | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
6J80
 
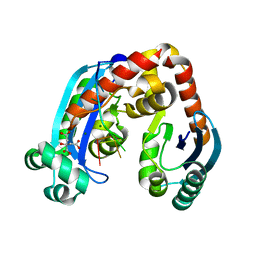 | | Human mitochondrial Oligoribonuclease in complex with poly-dT DNA | | Descriptor: | CITRIC ACID, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Chu, L.Y, Agrawal, S, Yuan, H.S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Structural insights into nanoRNA degradation by human Rexo2.
Rna, 25, 2019
|
|
6N5P
 
 | | Structure of Human pir-miRNA-340 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5T
 
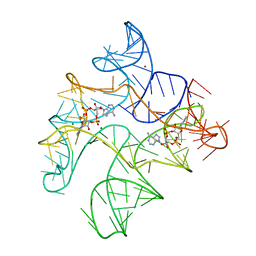 | | Structure of Human pir-miRNA-378a Apical Loop Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5K
 
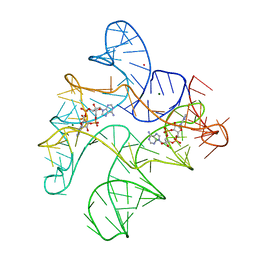 | | Structure of Human pir-miRNA-449c Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5O
 
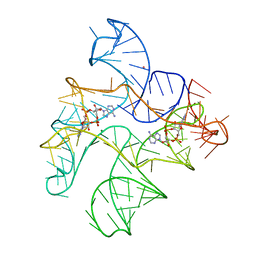 | | Structure of Human pir-miRNA-202 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5S
 
 | | Structure of Human pir-miRNA-320b-2 Apical Loop and One-base-pair Stem Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5N
 
 | | Structure of Human pir-miRNA-208a Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5Q
 
 | | Structure of Human pir-miRNA-378a Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.946 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
7N3O
 
 | | Cryo-EM structure of the Cas12k-sgRNA complex | | Descriptor: | Cas12k, Single guide RNA | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
7N3P
 
 | | Cryo-EM structure of the Cas12k-sgRNA-dsDNA complex | | Descriptor: | Cas12k, DNA (5'-D(*CP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*AP*AP*CP*CP*GP*AP*GP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*CP*TP*CP*GP*GP*TP*T)-3'), ... | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
1FYJ
 
 | | SOLUTION STRUCTURE OF MULTI-FUNCTIONAL PEPTIDE MOTIF-1 PRESENT IN HUMAN GLUTAMYL-PROLYL TRNA SYNTHETASE (EPRS). | | Descriptor: | MULTIFUNCTIONAL AMINOACYL-TRNA SYNTHETASE | | Authors: | Jeong, E.-J, Hwang, G.-S, Kim, K.H, Kim, M.J, Kim, S, Kim, K.-S. | | Deposit date: | 2000-10-02 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of multifunctional peptide motifs in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats.
Biochemistry, 39, 2000
|
|
3HFY
 
 | | Mutant of tRNA-guanine transglycosylase (K52M) | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ritschel, T, Klebe, G. | | Deposit date: | 2009-05-13 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Integrative Approach Combining Noncovalent Mass Spectrometry, Enzyme Kinetics and X-ray Crystallography to Decipher Tgt Protein-Protein and Protein-RNA Interaction
J.Mol.Biol., 393, 2009
|
|
2F4U
 
 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-((1R,2S,3R,4R,5S)-5-AMINO-4-[(2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-{2-[(3-AMINOPROPYL)AMINO]ETHOXY}-3-HYDROXYCYCLOHEXYL)-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
