2J1A
 
 | |
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | Descriptor: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
2QUO
 
 | |
1PFO
 
 | | PERFRINGOLYSIN O | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form.
Cell(Cambridge,Mass.), 89, 1997
|
|
5TSP
 
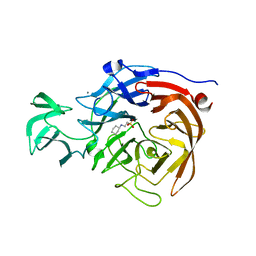 | | Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Sialidase | | Authors: | Lee, Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Kang, J.Y, Jin, M.S, Ryu, Y.B, Park, K.H, Eom, S.H. | | Deposit date: | 2016-10-31 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase in complex with a non-carbohydrate-based inhibitor, 2-(cyclohexylamino)ethanesulfonic acid
Biochem. Biophys. Res. Commun., 486, 2017
|
|
2WGW
 
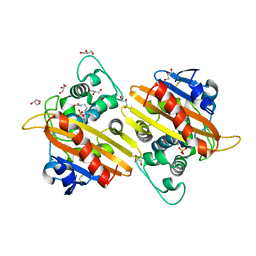 | | Crystal structure of the OXA-10 V117T mutant at pH 8.0 | | Descriptor: | BETA-LACTAMASE OXA-10, GLYCEROL, SULFATE ION | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2WGV
 
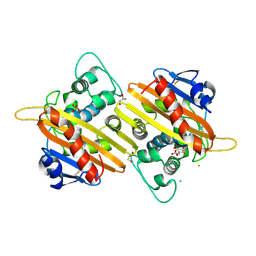 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2WKH
 
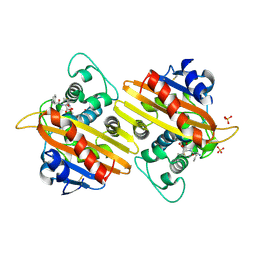 | | Crystal structure of the acyl-enzyme OXA-10 K70C-Ampicillin at pH 7 | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, BETA-LACTAMASE OXA-10, SULFATE ION | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2WKI
 
 | | Crystal structure of the OXA-10 K70C mutant at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GLYCEROL, ... | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
5WX9
 
 | | Crystal Structure of AtERF96 with GCC-box | | Descriptor: | Ethylene-responsive transcription factor ERF096, GCC-box motif | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2017-01-06 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box.
Plant Mol.Biol., 104, 2020
|
|
1R5O
 
 | | crystal structure analysis of sup35 complexed with GMPPNP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-11 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5B
 
 | | Crystal structure analysis of sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5N
 
 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
8HFC
 
 | | Cryo-EM structure of yeast Erf2/Erf4 complex | | Descriptor: | PALMITIC ACID, Palmitoyltransferase ERF2, Ras modification protein ERF4, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Yang, A, Liu, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6D7A
 
 | | Structure of T. gondii PLP1 beta-rich domain | | Descriptor: | Perforin-like protein 1, SODIUM ION | | Authors: | Guerra, A.J, Koropatkin, N.M, Wawrzak, Z, Bahr, C.M.E, Carruthers, V.B. | | Deposit date: | 2018-04-24 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural basis of Toxoplasma gondii perforin-like protein 1 membrane interaction and activity during egress.
PLoS Pathog., 14, 2018
|
|
3KZX
 
 | |
3D30
 
 | | Structure of an expansin like protein from Bacillus Subtilis at 1.9A resolution | | Descriptor: | Expansin like protein, FORMIC ACID, GLYCEROL | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D2Z
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3D2Y
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
6CS2
 
 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS1
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRX
 
 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRW
 
 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
5UOT
 
 | |
