9J8G
 
 | | mouse zbp1 hybrid complex | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*CP*A)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*G)-3'), Z-DNA-binding protein 1 | | Authors: | Gao, A.M, Zhoiu, C. | | Deposit date: | 2024-08-21 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZBP1 senses spliceosome stress through Z-RNA:DNA hybrid recognition.
Mol.Cell, 85, 2025
|
|
9J89
 
 | | zbp1 nucleic acid complex | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*CP*A)-3'), MAGNESIUM ION, RNA (5'-R(P*UP*GP*CP*GP*UP*G)-3'), ... | | Authors: | Gao, A.M, Zhou, C. | | Deposit date: | 2024-08-20 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ZBP1 senses spliceosome stress through Z-RNA:DNA hybrid recognition.
Mol.Cell, 85, 2025
|
|
2IG3
 
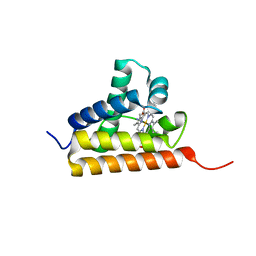 | | Crystal structure of group III truncated hemoglobin from Campylobacter jejuni | | Descriptor: | ACETATE ION, CYANIDE ION, Group III truncated haemoglobin, ... | | Authors: | Nardini, M, Pesce, A, Labarre, M, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural determinants in the group III truncated hemoglobin from Campylobacter jejuni.
J.Biol.Chem., 281, 2006
|
|
2O3B
 
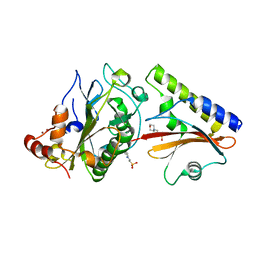 | | Crystal structure complex of Nuclease A (NucA) with intra-cellular inhibitor NuiA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The nuclease a-inhibitor complex is characterized by a novel metal ion bridge.
J.Biol.Chem., 282, 2007
|
|
4YOO
 
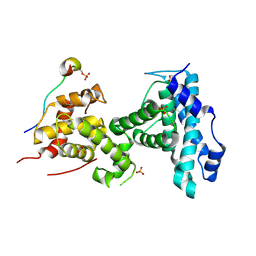 | | p107 pocket domain in complex with LIN52 P29A peptide | | Descriptor: | LIN52 peptide, Retinoblastoma-like protein 1,Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Guiley, K.Z, Liban, T.J, Felthousen, J.G, Ramanan, P, Tripathi, S, Litovchick, L, Rubin, S.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanisms of DREAM complex assembly and regulation.
Genes Dev., 29, 2015
|
|
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6E6C
 
 | | HRAS G13D bound to GppNHp (H13GNP) | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6E6H
 
 | | NRAS G13D bound to GppNHp (N13GNP) | | Descriptor: | GTPase NRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6YXX
 
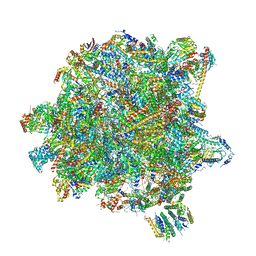 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6Z3T
 
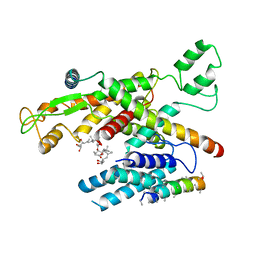 | | Structure of canine Sec61 inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Gerard, S.F, Higgins, M.K. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Inhibited State of the Sec Translocon.
Mol.Cell, 79, 2020
|
|
6XX0
 
 | | Crystal structure of NEMO in complex with Ubv-LIN | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b, ... | | Authors: | Akutsu, M, Skenderovic, A, Garcia-Pardo, J, Maculins, T, Dikic, I. | | Deposit date: | 2020-01-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
8QCF
 
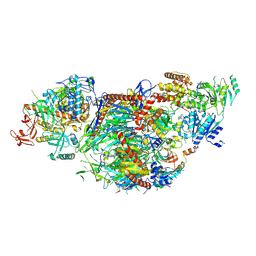 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
4AM3
 
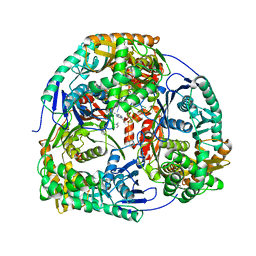 | | Crystal structure of C. crescentus PNPase bound to RNA | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
6XSK
 
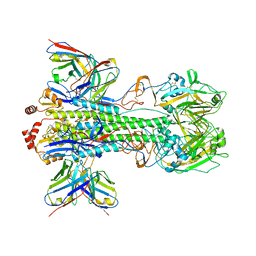 | | Cryo-EM Structure of Vaccine-Elicited Rhesus Antibody 789-203-3C12 in Complex with Stabilized SI06 (A/Solomon Islands/3/06) Influenza Hemagglutinin Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 789-203-3C12 Fab Heavy Chain, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses.
Immunity, 55, 2022
|
|
8QB6
 
 | | Dispersin from Terribacillus saccharophilus DispTs2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, Dispersin | | Authors: | Males, A, Moroz, O.V, Blagova, E, Munch, A, Hansen, G.H, Johansen, A.H, Ostergaard, L.H, Segura, D.R, Eddenden, A, Due, A.V, Gudmand, M, Salomon, J, Sorensen, S.R, Cairo, J.L.F, Pache, R.A, Vejborg, R.M, Davies, G.J, Wilson, K.S. | | Deposit date: | 2023-08-24 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Expansion of the diversity of dispersin scaffolds.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4NK2
 
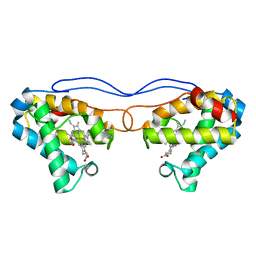 | | Crystal structure of Hell's gate globin IV | | Descriptor: | Hemoglobin-like protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jamil, F. | | Deposit date: | 2013-11-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of truncated haemoglobin from an extremely thermophilic and acidophilic bacterium.
J.Biochem., 156, 2014
|
|
8Q9T
 
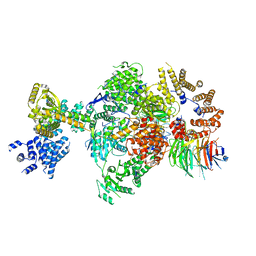 | | CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
4MBA
 
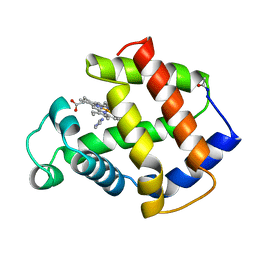 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | IMIDAZOLE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
8CZH
 
 | | Human BAK in complex with the dM2 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, DM2 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
8CZF
 
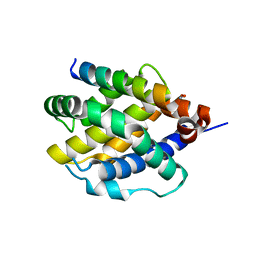 | | Human BAK in complex with the dF2 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, DF2 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
8CZG
 
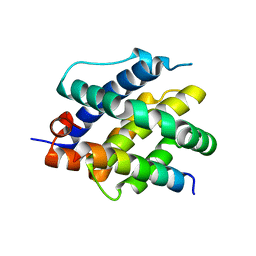 | | Human BAK in complex with the dF3 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, dF3 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
4NK1
 
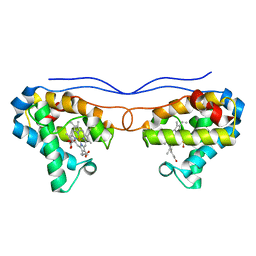 | | Crystal structure of phosphate-bound Hell's gate globin IV | | Descriptor: | Hemoglobin-like protein, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jamil, F. | | Deposit date: | 2013-11-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of truncated haemoglobin from an extremely thermophilic and acidophilic bacterium.
J.Biochem., 156, 2014
|
|
4QKO
 
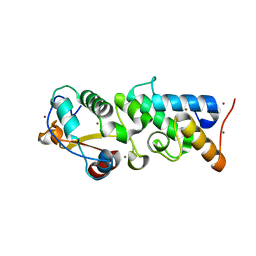 | | The Crystal Structure of the Pyocin S2 Nuclease Domain, Immunity Protein Complex at 1.8 Angstroms | | Descriptor: | BROMIDE ION, MAGNESIUM ION, Pyocin-S2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, C.J, Walker, D. | | Deposit date: | 2014-06-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into pyocin S2
To be Published
|
|
7OMB
 
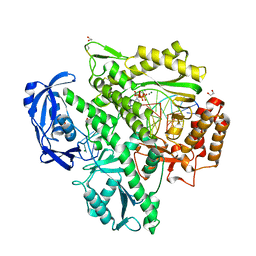 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OM3
 
 | | Crystal structure of KOD DNA Polymerase in a binary complex with Hypoxanthine containing template | | Descriptor: | 1,2-ETHANEDIOL, 21nt Template, BROMIDE ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
