6LMS
 
 | |
6LR2
 
 | |
6I7Z
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 32.8 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6I8E
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 65.6 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6S1E
 
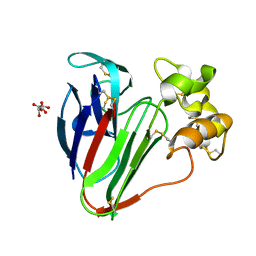 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
2BZA
 
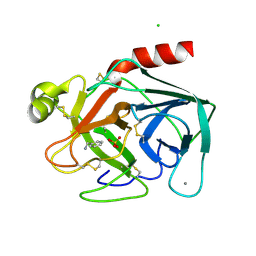 | | BOVINE PANCREAS BETA-TRYPSIN IN COMPLEX WITH BENZYLAMINE | | Descriptor: | BENZYLAMINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ota, N, Stroupe, C, Ferreira-Da-Silva, J.M.S, Shah, S.S, Mares-Guia, M, Brunger, A.T. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Boltzmann thermodynamic integration (NBTI) for macromolecular systems: relative free energy of binding of trypsin to benzamidine and benzylamine.
Proteins, 37, 1999
|
|
2C8P
 
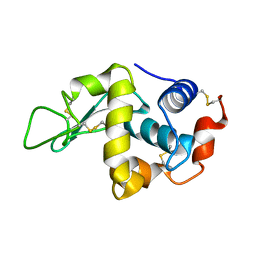 | | lysozyme (60sec) and UV laser excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6I57
 
 | |
2ZCE
 
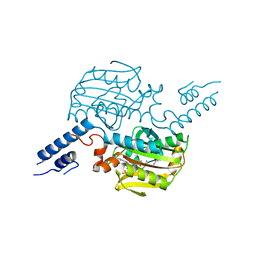 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with pyrrolysine and an ATP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYRROLYSINE, ... | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies on Multiple Conformational States of Active-site Loops in Pyrrolysyl-tRNA Synthetase
J.Mol.Biol., 378, 2008
|
|
6IJW
 
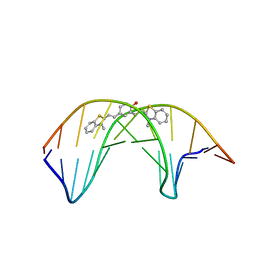 | |
2C8Q
 
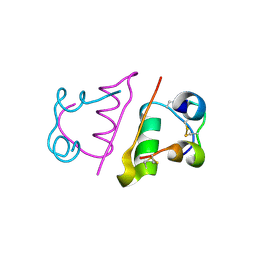 | | insuline(1sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6LUK
 
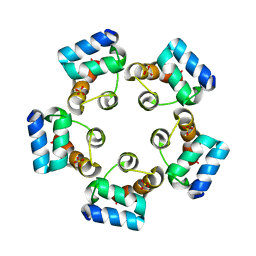 | |
4OCA
 
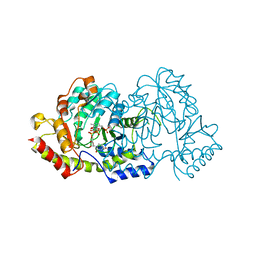 | | Cryatal structure of ArnB K188A complexted with PLP and UDP-Ara4N | | Descriptor: | (2R,3R,4S,5S)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]tetrahydro-2H-pyr an-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | Authors: | Sousa, M.C, Lee, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria.
Biochemistry, 53, 2014
|
|
6RWG
 
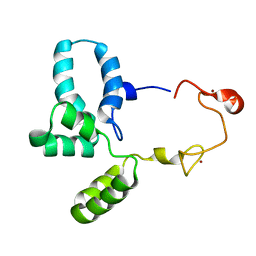 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
6S0N
 
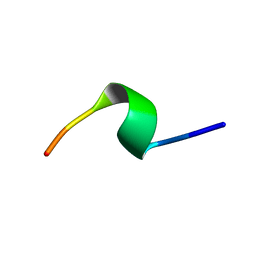 | | A9 peptide derived from Herceptin fab binding region | | Descriptor: | GLN-ASP-VAL-ASN-THR-ALA-VAL-ALA-TRP | | Authors: | De Luca, S, Verdoliva, V, Saviano, M, Fattorusso, R, Diana, D. | | Deposit date: | 2019-06-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPR and NMR characterization of the molecular interaction between A9 peptide and a model system of HER2 receptor: A fragment approach for selecting peptide structures specific for their target.
J.Pept.Sci., 26, 2020
|
|
6SAI
 
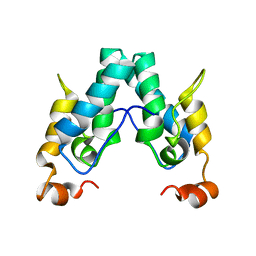 | |
2DV0
 
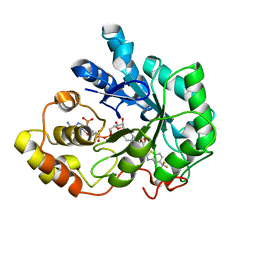 | | Human Aldose Reductase complexed with zopolrestat after 6 days soaking(6days_soaked_2) | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-07-28 | | Release date: | 2006-10-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions
J.Mol.Biol., 363, 2006
|
|
6E3C
 
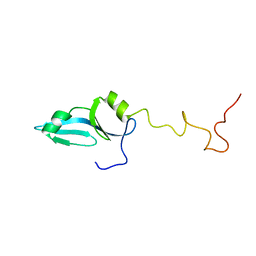 | |
1EX8
 
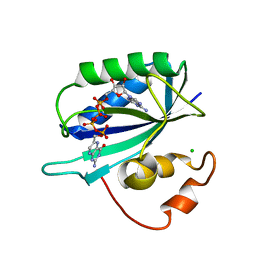 | | CRYSTAL STRUCTURE OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE COMPLEXED WITH HP4A, THE TWO-SUBSTRATE-MIMICKING INHIBITOR | | Descriptor: | 6-(ADENOSINE TETRAPHOSPHATE-METHYL)-7,8-DIHYDROPTERIN, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-05-01 | | Release date: | 2001-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: synthesis and biochemical and crystallographic studies.
J.Med.Chem., 44, 2001
|
|
4ORB
 
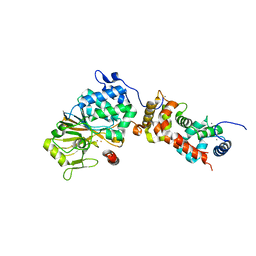 | | Crystal structure of mouse calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
3A5X
 
 | |
6IWS
 
 | | Solution structure of the J-domain of Tid1, a Mitochondrial Hsp40/DnaJ Protein | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Sim, D.W, Jo, K.S, Won, H.S, Kim, J.H. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the J-domain of Tid1, a Mitochondrial Hsp40/DnaJ Protein
To Be Published
|
|
2DUX
 
 | | Crystal structure of human Aldose Reductase complexed with zopolrestat after 3 days soaking (3days_soaked_1) | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-07-27 | | Release date: | 2006-10-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions
J.Mol.Biol., 363, 2006
|
|
6LXG
 
 | | NMR solution structure of regulatory ACT domain of the Mycobacterium tuberculosis Rel protein | | Descriptor: | GTP pyrophosphokinase | | Authors: | Shin, J, Singal, B, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2020-02-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
Febs J., 288, 2021
|
|
6LKF
 
 | |
