2AFI
 
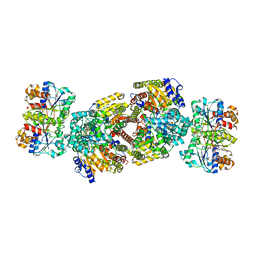 | | Crystal Structure of MgADP bound Av2-Av1 Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
1MV0
 
 | |
1N1Z
 
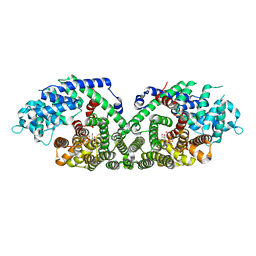 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and pyrophosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2A6U
 
 | |
7QTR
 
 | | GB1 in mammalian cells, 50 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
6I8J
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 131.2 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6I8Q
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 117.6 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
3GUB
 
 | | Crystal structure of DAPKL93G complexed with N6-(2-Phenylethyl)adenosine | | Descriptor: | 9-alpha-L-lyxofuranosyl-N-(2-phenylethyl)-9H-purin-6-amine, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-29 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant complexed with N6-modified adenosine analogs.
To be Published
|
|
6QTF
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, major conformer | | Descriptor: | DCY-LEU-GLY-ALA-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYU
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A | | Descriptor: | PHE-DHA-DAL-LEU-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6R1V
 
 | |
6QYR
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, minor conformer | | Descriptor: | DAL-LEU-GLY-CYS-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QM1
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B (Lan8,11) analogue | | Descriptor: | DAL-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-01 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6R95
 
 | |
6R5G
 
 | |
3GU8
 
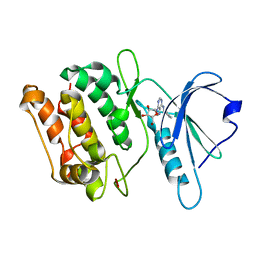 | | Crystal structure of DAPKL93G with N6-cyclopentyladenosine | | Descriptor: | Death-associated protein kinase 1, N6-cyclopentyladenosine | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant with N6-modified adenosine analogs.
To be Published
|
|
6I1O
 
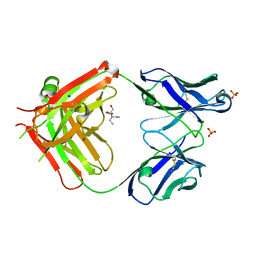 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
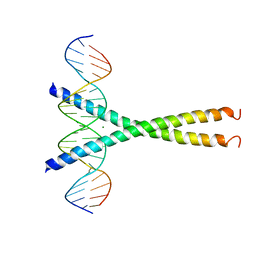
1DH3
 
 | |
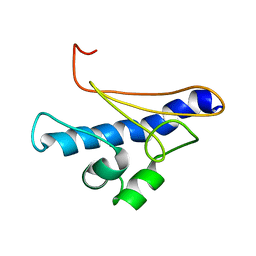
6I2O
 
 | |
6LCI
 
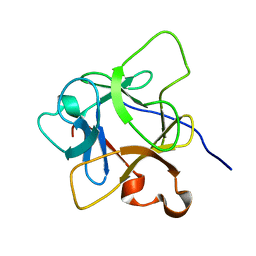 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
6L8M
 
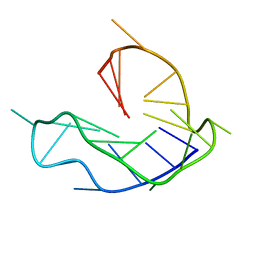 | | WNT DNA promoter mutant G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*CP*AP*CP*CP*GP*GP*GP*CP*AP*GP*TP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Wang, Z.F, Li, M.H, Chu, I.T, Winnerdy, F.R, Phan, A.T, Chang, T.C. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter.
Nucleic Acids Res., 48, 2020
|
|
6L7K
 
 | | solution structure of hIFABP V60C/Y70C variant. | | Descriptor: | Fatty acid-binding protein, intestinal | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.
Biophys.J., 118, 2020
|
|
2HFQ
 
 | | NMR structure of protein NE1680 from Nitrosomonas europaea: Northeast Structural Genomics Consortium target NeT5 | | Descriptor: | Hypothetical protein | | Authors: | Cort, J.R, Yee, A.A, Yim, V, Lukin, J, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-25 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein NE1680 from Nitrosomonas europaea: Northeast Structural Genomics Consortium target NeT5
TO BE PUBLISHED
|
|
2HQW
 
 | | Crystal Structure of Ca2+/Calmodulin bound to NMDA Receptor NR1C1 peptide | | Descriptor: | CALCIUM ION, Calmodulin, Glutamate NMDA receptor subunit zeta 1 | | Authors: | Akyol, Z, Gakhar, L, Sorensen, B.R, Hell, J.H, Shea, M.A. | | Deposit date: | 2006-07-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The NMDA Receptor NR1 C1 Region Bound to Calmodulin: Structural Insights into Functional Differences between Homologous Domains.
Structure, 15, 2007
|
|
