3BOP
 
 | | Structure of mouse beta-neurexin 2D4 | | Descriptor: | beta-Neurexin 2D4 | | Authors: | Koehnke, J, Jin, X, Shapiro, L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of beta-Neurexin 1 and beta-Neurexin 2 Ectodomains and Dynamics of Splice Insertion Sequence 4.
Structure, 16, 2008
|
|
3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
2A4C
 
 | | Crystal structure of mouse cadherin-11 EC1 | | Descriptor: | Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
3BZ4
 
 | | Crystal structure of Fab F22-4 in complex with a Shigella flexneri 2a O-Ag decasaccharide | | Descriptor: | Fab F22-4 heavy chain, Fab F22-4 light chain, PALLADIUM ION, ... | | Authors: | Saul, F.A, Vulliez-le-Normand, B, Bentley, G.A. | | Deposit date: | 2008-01-17 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of synthetic O-antigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C0W
 
 | | I-SceI in complex with a bottom nicked DNA substrate | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DAP*DAP*DCP*DAP*DGP*DGP*DGP*DTP*DAP*DAP*DTP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DT)-3'), ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway.
Nucleic Acids Res., 36, 2008
|
|
3C23
 
 | | Structure of a bacterial DNA damage sensor protein with non-reactive Ligand | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3BKM
 
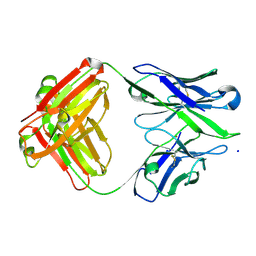 | | Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa, ... | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BVW
 
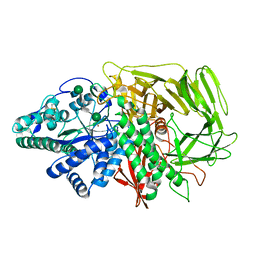 | |
3BYN
 
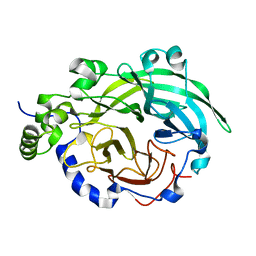 | |
3BYK
 
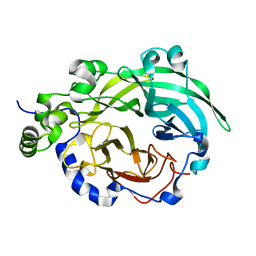 | |
3C5S
 
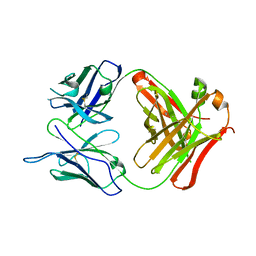 | |
3C1Z
 
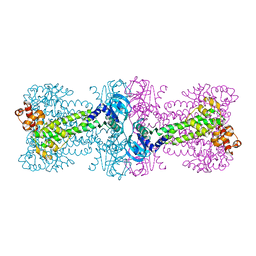 | |
3C7J
 
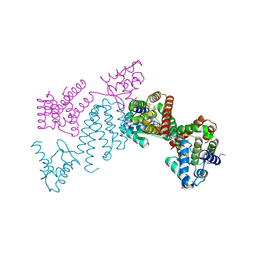 | | Crystal structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | NICKEL (II) ION, Transcriptional regulator, GntR family | | Authors: | Nocek, B, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-07 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
3C9M
 
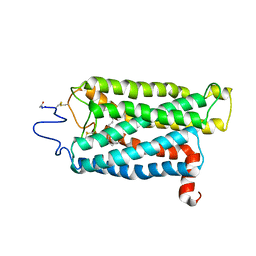 | |
3BNU
 
 | | Crystal structure of polyamine oxidase FMS1 from Saccharomyces cerevisiae in complex with bis-(3S,3'S)-methylated spermine | | Descriptor: | (3S,3'S)-N~1~,N~1~'-butane-1,4-diyldibutane-1,3-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the substrate stereospecificity of FMS1.
To Be Published
|
|
6RBJ
 
 | | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, CHLORIDE ION, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
3PRU
 
 | | Crystal Structure of Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 1 (fragment 14-158) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR182A | | Descriptor: | CHLORIDE ION, Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, ... | | Authors: | Kuzin, A, Su, M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR182A
To be Published
|
|
3Q0Z
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Syntheses and initial evaluation of a series of indolo-fused heterocyclic inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q3L
 
 | | The neutron crystallographic structure of inorganic pyrophosphatase from Thermococcus thioreducens | | Descriptor: | CALCIUM ION, Tt-IPPase | | Authors: | Hughes, R.C, Coates, L, Blakeley, M.P, Tomanicek, S.J, Meehan, E.J, Garcia-Ruiz, J.M, Ng, J.D. | | Deposit date: | 2010-12-22 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (2.5 Å) | | Cite: | Inorganic pyrophosphatase crystals from Thermococcus thioreducens for X-ray and neutron diffraction.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3Q4G
 
 | | Structure of NAD synthetase from Vibrio cholerae | | Descriptor: | CALCIUM ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of NAD synthetase from Vibrio cholerae
TO BE PUBLISHED
|
|
6RP7
 
 | | The crystal structure of a complex between the LlFpg protein, a THF-DNA and an inhibitor | | Descriptor: | 6,7-dimethyl-2-sulfanylidene-1~{H}-pteridin-4-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Goffinont, S, Castaing, B. | | Deposit date: | 2019-05-14 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights.
Int J Mol Sci, 21, 2020
|
|
7P98
 
 | | Cyclohex-1-ene-1-carboxyl-CoA dehydrogenase in a substrate-free state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Short-chain acyl-CoA dehydrogenase | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
6RO2
 
 | | The crystal structure of a complex between the LlFpg protein, a THF-DNA and an inhibitor | | Descriptor: | 2-sulfanyl-1,9-dihydro-6H-purin-6-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Goffinont, S, Castaing, B. | | Deposit date: | 2019-05-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights.
Int J Mol Sci, 21, 2020
|
|
6RRQ
 
 | | Crystal structure of tyrosinase PvdP from Pseudomonas aeruginosa bound to copper | | Descriptor: | COPPER (II) ION, GLYCEROL, PvdP | | Authors: | Wibowo, J.P, Batista, F.A, van Oosterwijk, N, Groves, M.R, Dekker, F.J, Quax, W.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel mechanism of inhibition by phenylthiourea on PvdP, a tyrosinase synthesizing pyoverdine of Pseudomonas aeruginosa.
Int.J.Biol.Macromol., 146, 2020
|
|
3PLS
 
 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
