1SXY
 
 | | 1.07 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SXZ
 
 | | Reduced bovine superoxide dismutase at pH 5.0 complexed with azide | | Descriptor: | AZIDE ION, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Wilson, K.S, Mangani, S. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site
J.Biol.Inorg.Chem., 3, 1998
|
|
1SY0
 
 | | 1.15 A Crystal Structure of T121V Mutant of Nitrophorin 4 from Rhodnius Prolixus | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SY1
 
 | | 1.0 A Crystal Structure of T121V Mutant of Nitrophorin 4 Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SY2
 
 | | 1.0 A Crystal Structure of D129A/L130A Mutant of Nitrophorin 4 | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SY3
 
 | | 1.00 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SY4
 
 | | Refined solution structure of the S. cerevisiae U6 INTRAMOLECULAR STEM LOOP (ISL) RNA USING RESIDUAL DIPOLAR COUPLINGS (RDCS) | | Descriptor: | U6 Intramolecular Stem-Loop RNA | | Authors: | Reiter, N.J, Nikstad, L.J, Allman, A.M, Johnson, R.J, Butcher, S.E. | | Deposit date: | 2004-03-31 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the U6 RNA intramolecular stem-loop harboring an
S(P)-phosphorothioate modification.
RNA, 9, 2003
|
|
1SY6
 
 | | Crystal Structure of CD3gammaepsilon Heterodimer in Complex with OKT3 Fab Fragment | | Descriptor: | OKT3 Fab heavy chain, OKT3 Fab light chain, T-cell surface glycoprotein CD3 gamma/epsilon chain | | Authors: | Kjer-Nielsen, L, Dunstone, M.A, Kostenko, L, Ely, L.K, Beddoe, T, Misfud, N.A, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-05-25 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human T cell receptor CD3(epsilon)(gamma) heterodimer complexed to the therapeutic mAb OKT3.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SY7
 
 | | Crystal structure of the catalase-1 from Neurospora crassa, native structure at 1.75A resolution. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Diaz, A, Horjales, E, Rudino-Pinera, E, Arreola, R, Hansberg, W. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unusual Cys-Tyr covalent bond in a large catalase
J.Mol.Biol., 342, 2004
|
|
1SY8
 
 | | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spec and restrained molecular dynamics | | Descriptor: | 5'-D(P*TP*GP*AP*TP*CP*A)-3' | | Authors: | Barthwal, R, Awasthi, P, Narang, M, Sharma, U, Srivastava, N. | | Deposit date: | 2004-04-01 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spectroscopy and restrained molecular dynamics
J.STRUCT.BIOL., 148, 2004
|
|
1SY9
 
 | | Structure of calmodulin complexed with a fragment of the olfactory CNG channel | | Descriptor: | CALCIUM ION, CALMODULIN, Cyclic-nucleotide-gated olfactory channel | | Authors: | Contessa, G.M, Orsale, M, Melino, S, Torre, V, Paci, M, Desideri, A, Cicero, D.O. | | Deposit date: | 2004-04-01 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of calmodulin complexed with an olfactory CNG channel fragment and role of the central linker: residual dipolar couplings to evaluate calmodulin binding modes outside the kinase family.
J.Biomol.Nmr, 31, 2005
|
|
1SYB
 
 | | TRANSFER OF A BETA-TURN STRUCTURE TO A NEW PROTEIN CONTEXT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Hynes, T.R, Kautz, R.A, Goodman, M.A, Gill, J.F, Fox, R.O. | | Deposit date: | 1994-01-07 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of a beta-turn structure to a new protein context.
Nature, 339, 1989
|
|
1SYC
 
 | |
1SYD
 
 | | ENGINEERING ALTERNATIVE BETA-TURN TYPES IN STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Hynes, T.R, Hodel, A, Fox, R.O. | | Deposit date: | 1994-01-07 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering alternative beta-turn types in staphylococcal nuclease.
Biochemistry, 33, 1994
|
|
1SYE
 
 | |
1SYF
 
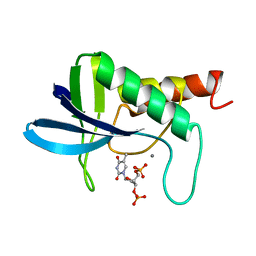 | | ENGINEERING ALTERNATIVE BETA-TURN TYPES IN STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Hynes, T.R, Hodel, A, Fox, R.O. | | Deposit date: | 1994-01-07 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering alternative beta-turn types in staphylococcal nuclease.
Biochemistry, 33, 1994
|
|
1SYG
 
 | |
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1SYI
 
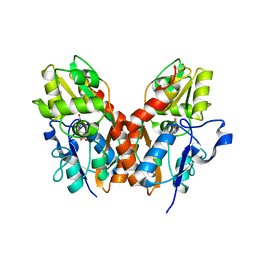 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1SYK
 
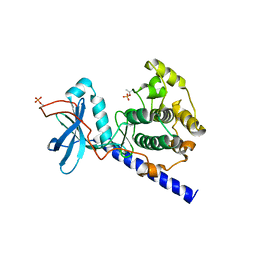 | | Crystal structure of E230Q mutant of cAMP-dependent protein kinase reveals unexpected apoenzyme conformation | | Descriptor: | cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Wu, J, Yang, J, Madhusudan, N, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the E230Q mutant of cAMP-dependent
protein kinase reveals an unexpected apoenzyme conformation and an
extended N-terminal A helix.
Protein Sci., 14, 2005
|
|
1SYL
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-04-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1SYM
 
 | |
1SYN
 
 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH BW1843U89 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Stroud, R.M. | | Deposit date: | 1995-09-19 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex of the anti-cancer therapeutic, BW1843U89, with thymidylate synthase at 2.0 A resolution: implications for a new mode of inhibition.
Structure, 4, 1996
|
|
1SYO
 
 | | N-terminal 3 domains of CI-MPR bound to mannose 6-phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose, GLYCEROL, ... | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2004-04-01 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminal carbohydrate recognition site of the cation-independent mannose 6-phosphate receptor
J.Biol.Chem., 279, 2004
|
|
1SYQ
 
 | |
