1L0E
 
 | |
1L3V
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 15 base pairs of duplex DNA following addition of dTTP, dATP, dCTP, and dGTP residues. | | Descriptor: | 5'-D(*GP*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*CP*GP*TP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MJ7
 
 | | Crystal Structure Of The Complex Of The Fab fragment of Esterolytic Antibody MS5-393 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS5-393, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1L1R
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
2JRL
 
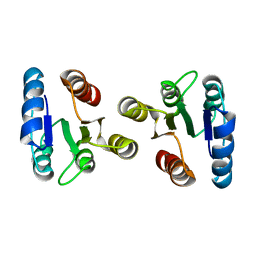 | | Solution structure of the beryllofluoride-activated NtrC4 receiver domain dimer | | Descriptor: | Transcriptional regulator (NtrC family) | | Authors: | Lee, C, Hong, E, Doucleff, M, Pelton, J.G, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Beryllofluoride-Activated NtrC4 Receiver Domain Dimer.
To be Published
|
|
1L4D
 
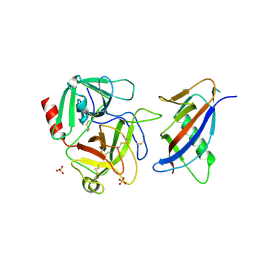 | | CRYSTAL STRUCTURE OF MICROPLASMINOGEN-STREPTOKINASE ALPHA DOMAIN COMPLEX | | Descriptor: | PLASMINOGEN, STREPTOKINASE, SULFATE ION | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-04 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation
PROTEIN ENG., 15, 2002
|
|
1L5N
 
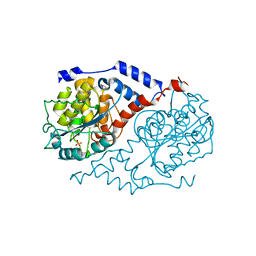 | | Crystal Structure of CobT complexed with imidazole | | Descriptor: | IMIDAZOLE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase
(CobD) enzyme from Salmonella enterica: the apo, substrate, and
product-aldimine complexes.
Biochemistry, 41, 2002
|
|
1L5J
 
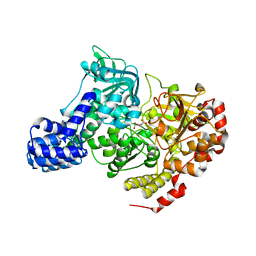 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
2JUR
 
 | |
2JWA
 
 | |
1L6P
 
 | |
1L6V
 
 | | STRUCTURE OF REDUCED BOVINE ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
2JYV
 
 | | Human Granulin F | | Descriptor: | Granulin-2 | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
1L8C
 
 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LC8
 
 | | Crystal Structure of L-Threonine-O-3-phosphate Decarboxylase from S. enterica complexed with its reaction intermediate | | Descriptor: | L-Threonine-O-3-Phosphate Decarboxylase, {3-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-PROPYL}-PHOSPHONIC ACID | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-04-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
Biochemistry, 41, 2002
|
|
2K36
 
 | | Structure ensemble Backbone and side-chain 1H, 13C, and 15N Chemical Shift Assignments, 1H-15N RDCs and 1H-1H nOe restraints for protein K7 from the Vaccinia virus | | Descriptor: | Protein K7 | | Authors: | Kalverda, A.P, Thompson, G.S, Vogel, A, Schr der, M, Bowie, A.G, Khan, A.R, Homans, S.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3.
J.Mol.Biol., 385, 2009
|
|
2JXN
 
 | | Solution Structure of S. cerevisiae PDCD5-like Protein Ymr074cp | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Uncharacterized protein YMR074C | | Authors: | Hong, J, Zhang, J, Liu, Z, Shi, Y, Wu, J. | | Deposit date: | 2007-11-23 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of S. cerevisiae PDCD5-like Protein Ymr074cp Determined by Heteronuclear NMR Spectroscopy
To be Published
|
|
1LCN
 
 | | Monoclinic hen egg white lysozyme, thiocyanate complex | | Descriptor: | PROTEIN (LYSOZYME), THIOCYANATE ION | | Authors: | Hamiaux, C, Prange, T, Ducruix, A, Vaney, M.C. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1LE1
 
 | |
2JYE
 
 | | Human Granulin A | | Descriptor: | Granulin A | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
1LGT
 
 | | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE (DHBD) COMPLEXED WITH 2'-Cl DIHYDROXYBIPHENYL (DHB) | | Descriptor: | 2'-CHLORO-BIPHENYL-2,3-DIOL, BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE, FE (II) ION | | Authors: | Dai, S, Bolin, J.T. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and analysis of a bottleneck in PCB biodegradation
Nat.Struct.Biol., 12, 2002
|
|
2JUQ
 
 | |
2K2W
 
 | | Second BRCT domain of NBS1 | | Descriptor: | Recombination and DNA repair protein | | Authors: | Xu, C, Cui, G, Botuyan, M, Mer, G. | | Deposit date: | 2008-04-14 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites.
J.Mol.Biol., 381, 2008
|
|
2JXY
 
 | | Solution structure of the hemopexin-like domain of MMP12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M. | | Deposit date: | 2007-12-01 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12
J.Am.Chem.Soc., 130, 2008
|
|
1LIQ
 
 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
