5K6O
 
 | |
7S6T
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit H33A | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
6WKS
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 in complex with RNA cap analogue (m7GpppA) and S-adenosylmethionine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, ADENOSINE, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Arya, S, Qi, S, Misra, A, Chan, S.-H. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of RNA cap modification by SARS-CoV-2.
Nat Commun, 11, 2020
|
|
6WUG
 
 | |
6WZ6
 
 | | Complex of mutant (K173M) of Pseudomonas 7A Glutaminase-Asparaginase with L-Glu at pH 5. Covalent acyl-enzyme intermediate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
7PZ3
 
 | | Structure of an LPMO at 5.37x10^3 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8E4Y
 
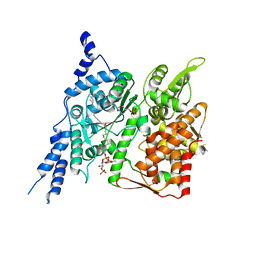 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase 1, mitochondrial, ... | | Authors: | Johnson, Z.L, Wasilko, D.J, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | Authors: | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2023-11-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 32, 2024
|
|
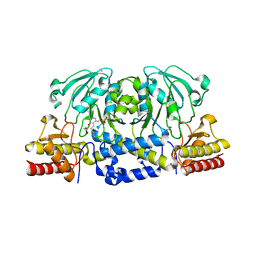
6XDK
 
 | |
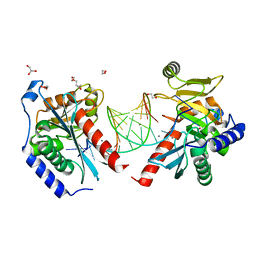
6OZQ
 
 | |
6PK2
 
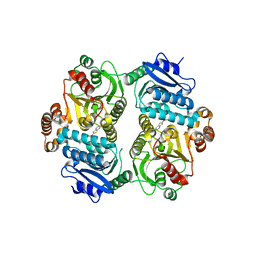 | | CRYSTAL STRUCTURE OF THE CARBOXYLTRANSFERASE SUBUNIT OF ACC (ACCD6) IN COMPLEX WITH INHIBITOR QUIZALOFOP-P derivative FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | 2-{4-[(6-fluoro-1,3-benzothiazol-2-yl)oxy]-2-hydroxyphenyl}-N-methylacetamide, Propionyl-CoA carboxylase subunit beta | | Authors: | Reddy, M.C.M, Nian, Z, Michele, T.C.B, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Elucidating the Inhibition and specificity of binding of
herbicidal aryloxyphenoxypropionates derivatives to Mycobacterium tuberculosis carboxyltransferase
domain of acetyl-coenzyme A(AccD6).
To Be Published
|
|
6QJY
 
 | |
6VWS
 
 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
6XWK
 
 | | Crystal structure of Phormidium rubidum phycocyanin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Sonani, R.R, Roszak, A.W, Cogdell, R.J, Madamwar, D, Liu, H, Gross, M.L, Blankenship, R.E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Revisiting high-resolution crystal structure of Phormidium rubidum phycocyanin.
Photosyn. Res., 144, 2020
|
|
6XUC
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6XW7
 
 | | Crystal structure of murine norovirus P domain in complex with Nanobody NB-5829 and glycochenodeoxycholate (GCDCA) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Kilic, T, Sabin, C, Hansman, G. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nanobody-Mediated Neutralization Reveals an Achilles Heel for Norovirus.
J.Virol., 94, 2020
|
|
8RDK
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MD420 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-pyridin-4-yl-1~{H}-pyrrolo[3,2-g]isoquinoline, Serine/threonine-protein kinase haspin, ... | | Authors: | Kraemer, A, Defois, M, Giraud, F, Moreau, P, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
6XK2
 
 | |
1FDG
 
 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (AMETANTRONE COMPLEX) | | Descriptor: | 1,4-BIS-[2-(2-HYDROXY-ETHYLAMINO)-ETHYLAMINO]-ANTHRAQUINONE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
9B3A
 
 | | filament of type 1 KD-mxyl miniature tau macrocycle derived from 4R tauopathic fold | | Descriptor: | 1,3-dimethylbenzene, AMINO GROUP, Microtubule-associated protein tau, ... | | Authors: | Xu, X, Angera, J.I, Rajewski, H.B, Jiang, W, Del Valle, R.J. | | Deposit date: | 2024-03-18 | | Release date: | 2025-03-26 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Macrocyclic beta-arch peptides that mimic the structure and function of disease-associated tau folds.
Nat.Chem., 17, 2025
|
|
9GY9
 
 | | The Cryo-EM structure of bacterial beta-1,3-glucan phosphorylase from family GH161 | | Descriptor: | beta-1,3-glucan phosphorylase | | Authors: | Cioci, G, Cooper, N, Ladeveze, S, Shayan, R. | | Deposit date: | 2024-10-02 | | Release date: | 2025-10-15 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | The Cryo-EM structure of bacterial beta-1,3-glucan phosphorylase from family GH161
To Be Published
|
|
8HAU
 
 | | NARROW LEAF 1 from Indica | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Zhang, S.J, He, Y.J, Wang, N, Zhang, W.J, Liu, C.M. | | Deposit date: | 2022-10-26 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | NARROW LEAF 1 from Indica
To Be Published
|
|
5YZ1
 
 | | Crystal structure of human Archease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein archease | | Authors: | Duan, S.Y, Li, J.X. | | Deposit date: | 2017-12-11 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of human archease, a key cofactor of tRNA splicing ligase complex.
Int.J.Biochem.Cell Biol., 122, 2020
|
|
9H5P
 
 | | Crystal structure of Thermoanaerobacterales bacterium monoamine oxidase in complex with putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Basile, L, Poli, C, Santema, L.L, Lesenciuc, R.C, Fraaije, M.W, Binda, C. | | Deposit date: | 2024-10-23 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Altering substrate specificity of a thermostable bacterial monoamine oxidase by structure-based mutagenesis.
Arch.Biochem.Biophys., 764, 2024
|
|
377D
 
 | | 5'-R(*CP*GP*UP*AP*CP*DG)-3' | | Descriptor: | RNA-DNA (5'-R(*CP*GP*UP*AP*CP*DG)-3') | | Authors: | Biswas, R, Mitra, S.N, Sundaralingam, M. | | Deposit date: | 1998-01-26 | | Release date: | 1998-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 A structure of a pyrimidine start alternating A-RNA hexamer r(CGUAC)dG.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
