8XXL
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8X36
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8XT0
 
 | | Cryo-EM structure of the human 55S mitoribosome with 5um Tigecycline | | Descriptor: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8VFL
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 290K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
9BZQ
 
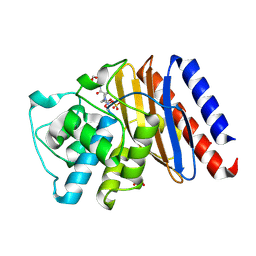 | | Structure of Class A Beta-lactamase from Bordetella bronchiseptica RB50 in a complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of Class A Beta-lactamase from Bordetella bronchiseptica RB50 in a complex with Avibactam
To Be Published
|
|
8VCH
 
 | |
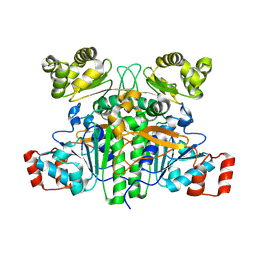
9BR6
 
 | |
9BNQ
 
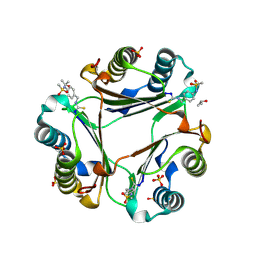 | | N-(4-(isothiocyanatomethyl)phenyl)methanesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-{[4-(methanesulfonamido)phenyl]methyl}methanethioamide, ... | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
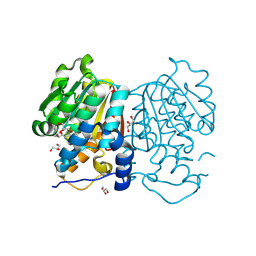
8Z8P
 
 | |
8VR1
 
 | |
8XFB
 
 | |
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
8VM2
 
 | | Crystal Structure of NRAS Q61K bound to GTP | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gebregiworgis, T, Chan, J.Y.L, Kuntz, D.A, Prive, G.G, Marshall, C.B, Ikura, M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of NRAS Q61K with a ligand-induced pocket near switch II.
Eur J Cell Biol, 103, 2024
|
|
8XZ3
 
 | |
8VR7
 
 | | crystal structure of the Pcryo_0619 N-acetyltransferase from Psychrobacter cryohalolentis K5 int he presence of acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
9EU9
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15i | | Descriptor: | (4-chloranyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
8WWX
 
 | |
8WXN
 
 | |
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
9BXA
 
 | |
8Y84
 
 | | Structure of the high affinity receptor fc(epsilon)ri TM | | Descriptor: | CHOLESTEROL HEMISUCCINATE, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure of the high affinity receptor fc(epsilon)ri TM
To Be Published
|
|
8YJM
 
 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and a single chain H2B-H2A chimera | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2B 1/2/3/4/6,Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
9ENI
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with L-glutamine | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLUTAMINE, L-amino acid oxidase 4, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum L-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum
to be published
|
|
8WZV
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 1 microsecond after photoexcitation with 40 microJoules laser intensity at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8VLC
 
 | | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, HARLDQ motif MBL-fold protein, SULFATE ION | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-11 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2
To Be Published
|
|
