7JSF
 
 | | Adeno-Associated Virus Helicase domain Heptamer with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
4ODA
 
 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
7DV2
 
 | | Structure of Sulfolobus solfataricus SegB-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*AP*GP*AP*AP*GP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*CP*TP*TP*CP*TP*AP*CP*GP*TP*A)-3'), SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7UDI
 
 | |
2KEI
 
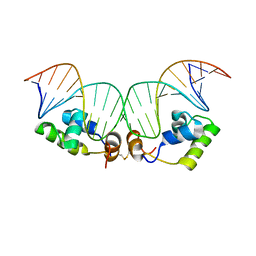 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
7X5L
 
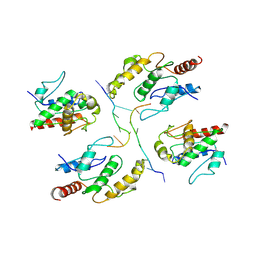 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*AP*TP*TP*AP*A)-3'), Flax rust resistance protein | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-04 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
4G4Q
 
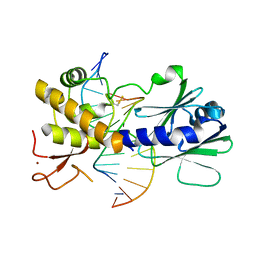 | | MutM containing F114A mutation bound to undamaged DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
3EPZ
 
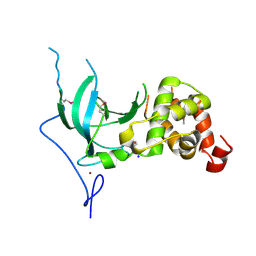 | | Structure of the replication foci-targeting sequence of human DNA cytosine methyltransferase DNMT1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, GLYCEROL, SODIUM ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1.
J.Biol.Chem., 286, 2011
|
|
3WPE
 
 | |
4M6F
 
 | |
2KEK
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
1DK3
 
 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
2KEJ
 
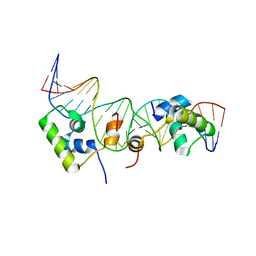 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
6OZI
 
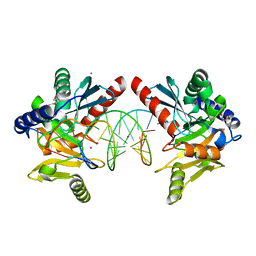 | |
6BEK
 
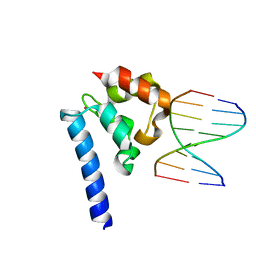 | |
6J4R
 
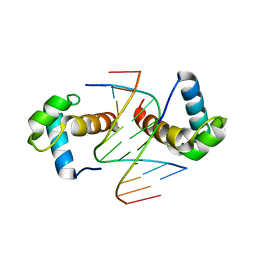 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
8SVA
 
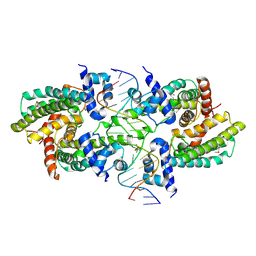 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
6J5B
 
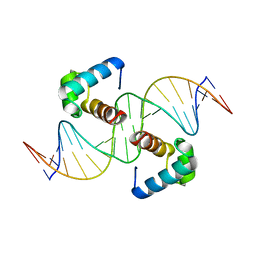 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
7WM3
 
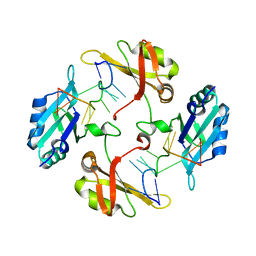 | | hnRNP A2/B1 RRMs in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Abula, A, Liu, Y, Guo, H, Li, T, Ji, X. | | Deposit date: | 2022-01-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2022
|
|
4B08
 
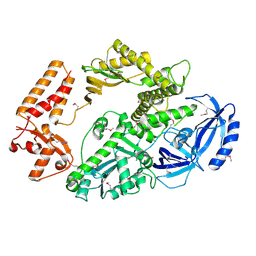 | | Yeast DNA polymerase alpha, Selenomethionine protein | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT A | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Mechanism for Priming DNA Synthesis by Yeast DNA Polymerase Alpha
Elife, 2, 2013
|
|
3K4X
 
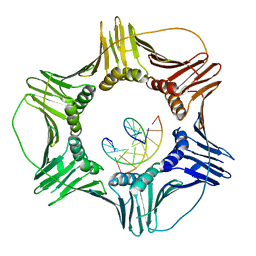 | | Eukaryotic Sliding Clamp PCNA Bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), Proliferating cell nuclear antigen | | Authors: | McNally, R, Kuriyan, J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Analysis of the role of PCNA-DNA contacts during clamp loading.
Bmc Struct.Biol., 10, 2010
|
|
1VT6
 
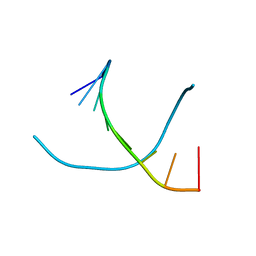 | |
6NHX
 
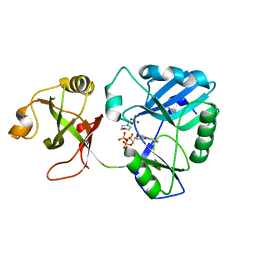 | | mycobacterial DNA ligase D complexed with ATP and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent DNA ligase | | Authors: | Shuman, S, Unciuleac, M, Goldgur, Y. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
J. Biol. Chem., 294, 2019
|
|
3ECP
 
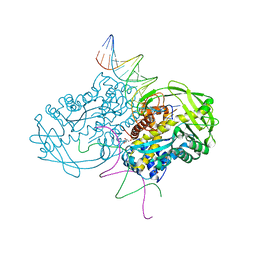 | |
3MGH
 
 | | Binary complex of a DNA polymerase lambda loop mutant | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Zhou, R.Z, Povirk, L.F, Kunkel, T. | | Deposit date: | 2010-04-06 | | Release date: | 2010-05-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Loop 1 modulates the fidelity of DNA polymerase lambda
Nucleic Acids Res., 38, 2010
|
|
