7WM4
 
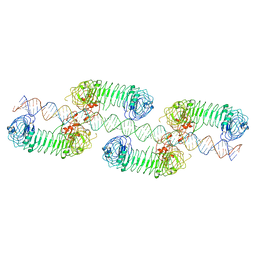 | | Cryo-EM structure of tetrameric TLR3 in complex with dsRNA (90 bp) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (81-MER), ... | | Authors: | Sakaniwa, K, Ohto, U, Shimizu, T. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TLR3 forms a laterally aligned multimeric complex along double-stranded RNA for efficient signal transduction.
Nat Commun, 14, 2023
|
|
4GOD
 
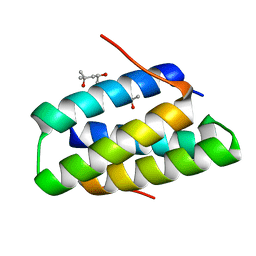 | | Crystal structure of the SGTA homodimerization domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Small glutamine-rich tetratricopeptide repeat-containing protein alpha | | Authors: | Chartron, J.W, VanderVelde, D.G, Clemons Jr, W.M. | | Deposit date: | 2012-08-19 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of the Sgt2/SGTA Dimerization Domain with the Get5/UBL4A UBL Domain Reveal an Interaction that Forms a Conserved Dynamic Interface.
Cell Rep, 2, 2012
|
|
4EP3
 
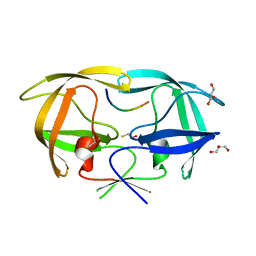 | | Crystal Structure of inactive single chain wild-type HIV-1 Protease in Complex with the substrate CA-p2 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
4OPO
 
 | |
4F3V
 
 | |
3G9C
 
 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-13 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
1Q2E
 
 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
4BAD
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxymethyltriazoledipicolinate complex at 1.35 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4BPA
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
5TRZ
 
 | | Crystal structure of MHC-I H2-KD complexed with peptides of Mycobacterial tuberculosis (YQSGLSIVM) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-10-27 | | Release date: | 2018-05-09 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | MHC-restricted Ag85B-specific CD8+T cells are enhanced by recombinant BCG prime and DNA boost immunization in mice.
Eur.J.Immunol., 2019
|
|
3GIP
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | Descriptor: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3G8T
 
 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
2I0Q
 
 | |
4BT9
 
 | |
4AJC
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, calcium(II) and adenine nucleotide phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-2'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4BTA
 
 | | CRYSTAL STRUCTURE OF THE PEPTIDE(PRO-PRO-GLY)3 BOUND COMPLEX OF N- TERMINAL DOMAIN AND PEPTIDE SUBSTRATE BINDING DOMAIN OF PROLYL-4 HYDROXYLASE (RESIDUES 1-244) TYPE I FROM HUMAN | | Descriptor: | PROLINE RICH PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Anantharajan, J, Koski, M.K, Pekkala, M, Wierenga, R.K. | | Deposit date: | 2013-06-14 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Structure, 21, 2013
|
|
2I6A
 
 | | Human Adenosine Kinase in Complex With 5'-Deoxy-5-Iodotubercidin | | Descriptor: | 7-(5-DEOXY-BETA-D-RIBOFURANOSYL)-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, Adenosine kinase | | Authors: | Muchmore, S.W. | | Deposit date: | 2006-08-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human adenosine kinase inhibitor complexes reveal two distinct binding modes.
J.Med.Chem., 49, 2006
|
|
1NMX
 
 | | Crystal structure of human thymidylate kinase with FLTMP and ADP | | Descriptor: | 3'-FLUORO-3'-DEOXYTHYMIDINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Segura-Pena, D, Meier, C, Veit, T, Monnerjahn, M, Konrad, M, Lavie, A. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of human thymidylate kinase in complex with prodrugs:
implications for the structure-based design of novel compounds
Biochemistry, 42, 2003
|
|
4BT8
 
 | |
3DKY
 
 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
4AJ3
 
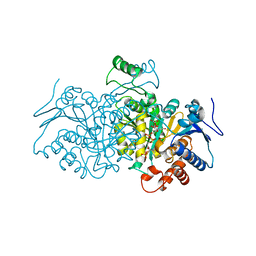 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and NADP - The pseudo-Michaelis complex | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, NADP ISOCITRATE DEHYDROGENASE, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJA
 
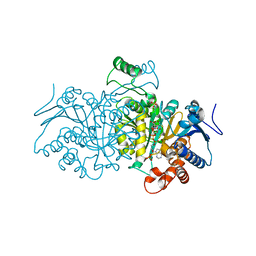 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
3ZVT
 
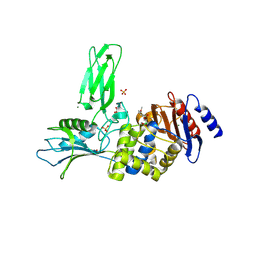 | | Unexpected tricovalent binding mode of boronic acids within the active site of a penicillin binding protein | | Descriptor: | 2,6 DIMETHOXYBENZAMIDOBORONIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Zervosen, A, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unexpected Tricovalent Binding Mode of Boronic Acids within the Active Site of a Penicillin- Binding Protein.
J.Am.Chem.Soc., 133, 2011
|
|
4J8D
 
 | |
4JA9
 
 | | Rat PP5 apo | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
