1OHA
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP and N-acetyl-L-glutamate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1OIS
 
 | | YEAST DNA TOPOISOMERASE I, N-TERMINAL FRAGMENT | | Descriptor: | DNA TOPOISOMERASE I | | Authors: | Lue, N, Sharma, A, Mondragon, A, Wang, J.C. | | Deposit date: | 1996-09-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 26 kDa yeast DNA topoisomerase I fragment: crystallographic structure and mechanistic implications.
Structure, 3, 1995
|
|
1OG5
 
 | | Structure of human cytochrome P450 CYP2C9 | | Descriptor: | CYTOCHROME P450 2C9, HEME C, S-WARFARIN | | Authors: | Williams, P.A, Cosme, J, Ward, A, Angove, H.C, Matak Vinkovic, D, Jhoti, H. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2C9 with Bound Warfarin
Nature, 424, 2003
|
|
1OFJ
 
 | |
1OI1
 
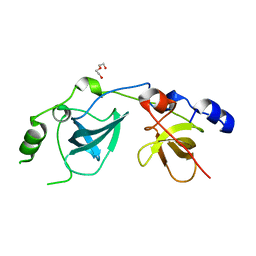 | | Crystal Structure of the MBT domains of Human SCML2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, SCML2 PROTEIN | | Authors: | Sathyamurthy, A, Allen, M.D, Murzin, A.G, Bycroft, M. | | Deposit date: | 2003-06-04 | | Release date: | 2003-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Malignant Brain Tumor (Mbt) Repeats in Sex Comb on Midleg-Like 2 (Scml2).
J.Biol.Chem., 278, 2003
|
|
1OGC
 
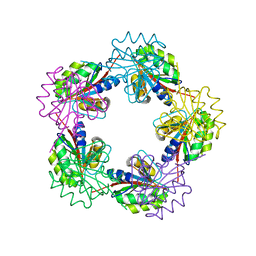 | | The Structure of Bacillus subtilis RbsD complexed with D-ribose | | Descriptor: | CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OI0
 
 | | CRYSTAL STRUCTURE OF AF2198, A JAB1/MPN DOMAIN PROTEIN FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | HYPOTHETICAL PROTEIN AF2198, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ZINC ION | | Authors: | Tran, H.J.T.T, Allen, M.D, Lowe, J, Bycroft, M. | | Deposit date: | 2003-06-04 | | Release date: | 2003-08-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Jab1/Mpn Domain and its Implications for Proteasome Function
Biochemistry, 42, 2003
|
|
1OGG
 
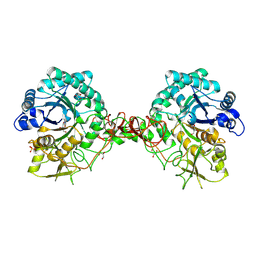 | | chitinase b from serratia marcescens mutant d142n in complex with inhibitor allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE B, ... | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-30 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
1OM0
 
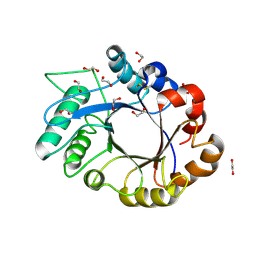 | | crystal structure of xylanase inhibitor protein (XIP-I) from wheat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Xylanase Inhibitor Protein I | | Authors: | Payan, F, Flatman, R, Porciero, S, Williamson, G, Juge, N, Roussel, A. | | Deposit date: | 2003-02-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson).
Biochem.J., 372, 2003
|
|
1OFT
 
 | |
1OO6
 
 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN23862 | | Descriptor: | 5-[BIS-2(CHLORO-ETHYL)-AMINO]-2,4-DINTRO-BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-03 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OT7
 
 | | Structural Basis for 3-deoxy-CDCA Binding and Activation of FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile Acid Receptor, ISO-URSODEOXYCHOLIC ACID, ... | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OPY
 
 | | KSI | | Descriptor: | DELTA5-3-KETOSTEROID IOSMERASE | | Authors: | Kim, S.-W, Cha, S.-S, Cho, H.-S, Kim, J.-S, Ha, N.-C, Cho, M.-J, Choi, K.-Y, Oh, B.-H. | | Deposit date: | 1997-05-23 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of delta5-3-ketosteroid isomerase with and without a reaction intermediate analogue.
Biochemistry, 36, 1997
|
|
1ORO
 
 | | A FLEXIBLE LOOP AT THE DIMER INTERFACE IS A PART OF THE ACTIVE SITE OF THE ADJACENT MONOMER OF ESCHERICHIA COLI OROTATE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | OROTATE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Henriksen, A, Aghajari, N, Jensen, K.F, Gajhede, M. | | Deposit date: | 1995-09-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase.
Biochemistry, 35, 1996
|
|
1OZF
 
 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactors | | Descriptor: | Acetolactate synthase, catabolic, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1OGD
 
 | | The Structure of Bacillus subtilis RbsD complexed with D-ribose | | Descriptor: | CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD, beta-D-ribopyranose | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1P1W
 
 | |
1OJM
 
 | |
1OKE
 
 | |
1ONA
 
 | | CO-CRYSTALS OF CONCANAVALIN A WITH METHYL-3,6-DI-O-(ALPHA-D-MANNOPYRANOSYL)-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Bouckaert, J, Maes, D, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1996-07-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structure of the complex between concanavalin A and methyl-3,6-di-O-(alpha-D-mannopyranosyl)-alpha-D-mannopyranoside reveals two binding modes.
J.Biol.Chem., 271, 1996
|
|
1P6I
 
 | | Rat neuronal NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(4S)-4-AMINO-5-[(2-AMINOETHYL)AMINO]PENTYL}-N'-NITROGUANIDINE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P7H
 
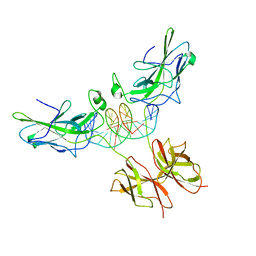 | | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*A)-3', 5'-D(*TP*TP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Giffin, M.J, Stroud, J.C, Bates, D.L, von Koenig, K.D, Hardin, J, Chen, L. | | Deposit date: | 2003-05-01 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Nat.Struct.Biol., 10, 2003
|
|
1OJZ
 
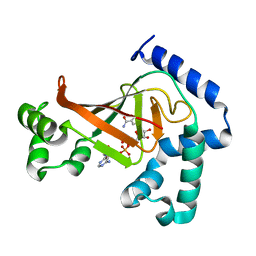 | | The crystal structure of C3stau2 from S. aureus with NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1OKB
 
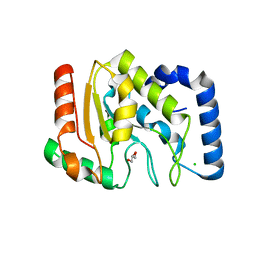 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2003-07-21 | | Release date: | 2004-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OOA
 
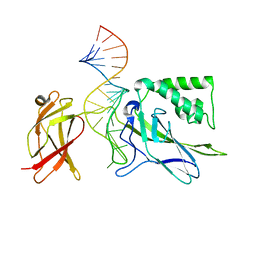 | | CRYSTAL STRUCTURE OF NF-kB(p50)2 COMPLEXED TO A HIGH-AFFINITY RNA APTAMER | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit, RNA aptamer | | Authors: | Huang, D.B, Vu, D, Cassiday, L.A, Zimmerman, J.M, Maher III, L.J, Ghosh, G. | | Deposit date: | 2003-03-03 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
