7D1U
 
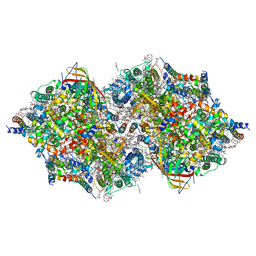 | | Cryo-EM Structure of PSII at 2.08 angstrom resolution | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kato, K, Miyazaki, N, Hamaguchi, T, Nakajima, Y, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams.
Commun Biol, 4, 2021
|
|
7DL9
 
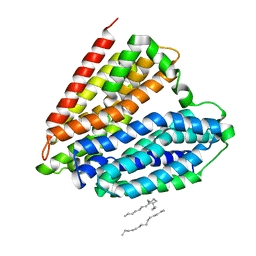 | | Crystal structure of nucleoside transporter NupG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Nucleoside permease NupG | | Authors: | Wang, C, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for substrate recognition by the bacterial nucleoside transporter NupG.
J.Biol.Chem., 296, 2021
|
|
7D80
 
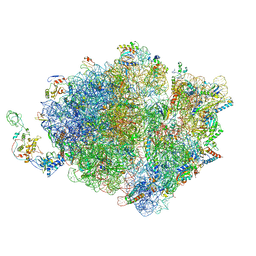 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
7D6Z
 
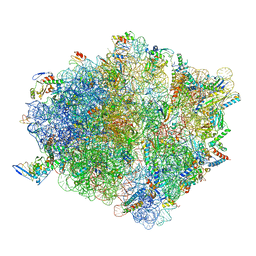 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase and trigger factor | | Descriptor: | 16S ribosomal rRNA, 23S ribosomal rRNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
2FOQ
 
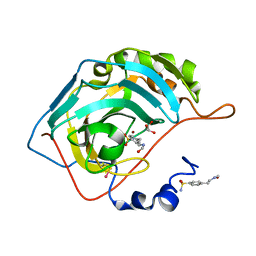 | |
7DLA
 
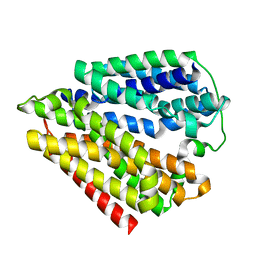 | |
8UDV
 
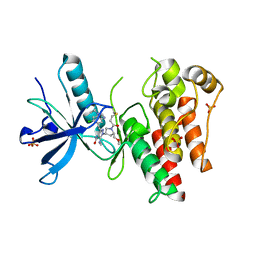 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
7D14
 
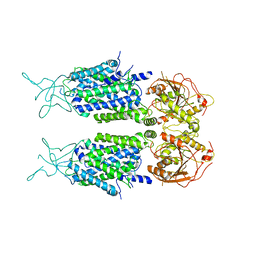 | | Mouse KCC2 | | Descriptor: | Solute carrier family 12 member 5 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
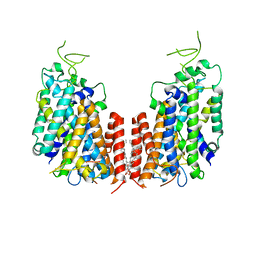 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
8U61
 
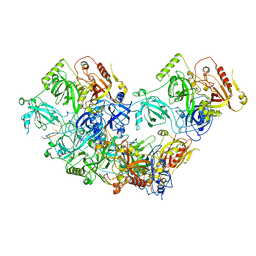 | | Human RADX tetramer bound to ssDNA | | Descriptor: | RPA-related protein RADX, dT25 DNA (25-MER) | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2FOU
 
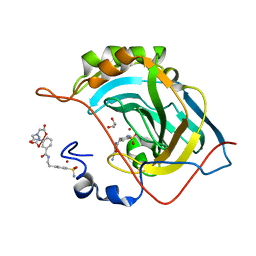 | | Human Carbonic Anhydrase II complexed with two-prong inhibitors | | Descriptor: | COPPER (II) ION, Carbonic Anhydrase II, GLYCEROL, ... | | Authors: | Jude, K.M, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
2FSE
 
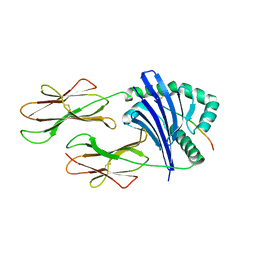 | | Crystallographic structure of a rheumatoid arthritis MHC susceptibility allele, HLA-DR1 (DRB1*0101), complexed with the immunodominant determinant of human type II collagen | | Descriptor: | Collagen alpha-1(II), H-2 class II histocompatibility antigen, E-K alpha chain, ... | | Authors: | Ivey, R.A, Rosloniec, E.F, Whittington, K.B, Kang, A.H, Park, H.W. | | Deposit date: | 2006-01-22 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Structure of a Rheumatoid Arthritis MHC Susceptibility Allele, HLA-DR1 (DRB1*0101), Complexed with the Immunodominant Determinant of Human Type II Collagen.
J.Immunol., 177, 2006
|
|
8TOQ
 
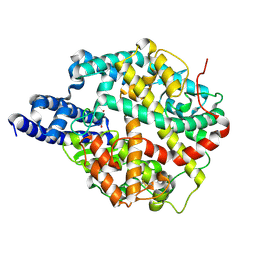 | | ACE2-peptide 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOS
 
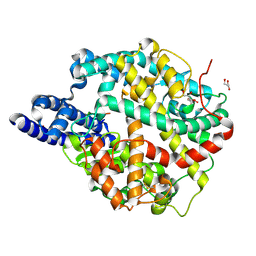 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TA5
 
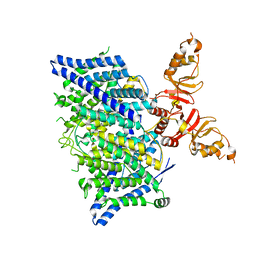 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
2FSA
 
 | |
8TA4
 
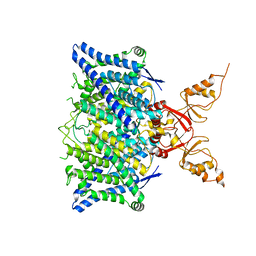 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
2FOV
 
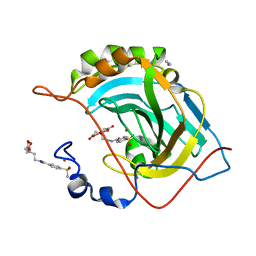 | | Human Carbonic Anhydrase II complexed with two-prong inhibitors | | Descriptor: | ACETONITRILE, COPPER (II) ION, Carbonic Anhydrase II, ... | | Authors: | Jude, K.M, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
8U18
 
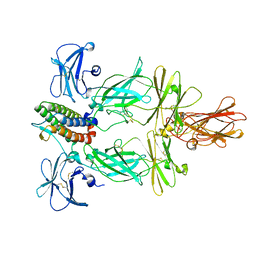 | | Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombopoietin, Thrombopoietin receptor,GCN4 isoform 1, ... | | Authors: | Sarson-Lawrence, K.S, Hardy, J.M, Leis, A, Babon, J.J, Kershaw, N.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin.
Nat Commun, 15, 2024
|
|
8TOR
 
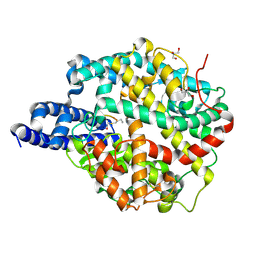 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
2FOY
 
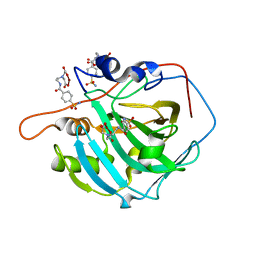 | | Human Carbonic Anhydrase I complexed with a two-prong inhibitor | | Descriptor: | Carbonic anhydrase 1, ZINC ION, {2,2'-[(2-{[4-(AMINOSULFONYL)BENZOYL]AMINO}ETHYL)IMINO]DIACETATO(2-)-KAPPAO}COPPER | | Authors: | Jude, K.M, Banerjee, A.L, Haldar, M.K, Manokaran, S, Roy, B, Mallik, S, Srivastava, D.K, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
2HFH
 
 | |
8TA2
 
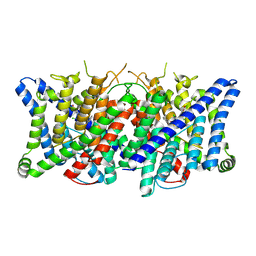 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
2HCO
 
 | |
8UDU
 
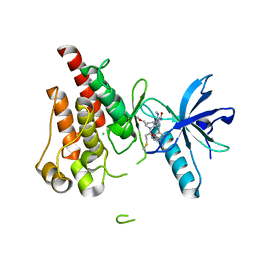 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
