5WLN
 
 | |
4OY3
 
 | |
2G0H
 
 | | Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities | | Descriptor: | N-[1-(4-FLUOROPHENYL)-3-(2-THIENYL)-1H-PYRAZOL-5-YL]-3,5-BIS(TRIFLUOROMETHYL)BENZENESULFONAMIDE, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Huang, C.F, Lin, Y.T, Hsu, J.T.A, Wu, S.Y. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Drug Design of a Novel Family of PPARgamma Partial Agonists: Virtual Screening, X-ray Crystallography, and in Vitro/in Vivo Biological Activities
J.Med.Chem., 49, 2006
|
|
8T3S
 
 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | Descriptor: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
4OYL
 
 | |
3LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS WITH PHOSPHOCHOLINE BOUND | | Descriptor: | LEUKOCIDIN F SUBUNIT, PHOSPHOCHOLINE | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
1PI7
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
3LO2
 
 | |
8SXJ
 
 | | CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab | | Descriptor: | CH235.12 Heavy Chain, CH235.12 Light Chain, Envelope glycoprotein gp160, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
2GEM
 
 | |
7KDP
 
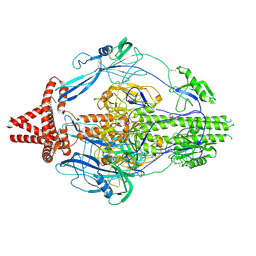 | | HCMV prefusion gB in complex with fusion inhibitor WAY-174865 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
7KDD
 
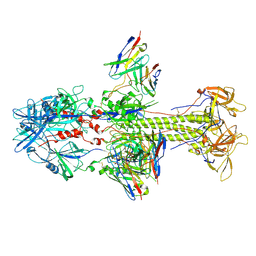 | | HCMV postfusion gB in complex with SM5-1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-08 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
2GH8
 
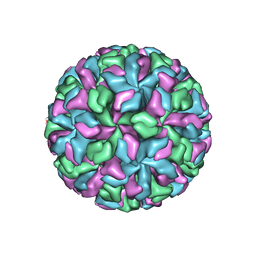 | | X-ray structure of a native calicivirus | | Descriptor: | Capsid protein | | Authors: | Chen, R. | | Deposit date: | 2006-03-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3LIN
 
 | | crystal structure of HTLV protease complexed with the inhibitor, KNI-10562 | | Descriptor: | N-[(2S,3S)-4-{(4R)-4-[(2,2-dimethylpropyl)carbamoyl]-5,5-dimethyl-1,3-thiazolidin-3-yl}-3-hydroxy-4-oxo-1-phenylbutan-2-yl]-N~2~-{(2S)-2-[(methoxycarbonyl)amino]-2-phenylacetyl}-3-methyl-L-valinamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
8CFA
 
 | |
3L81
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein, GLYCEROL | | Authors: | Mardones, G.A, Rojas, A.L, Burgos, P.V, Dasilva, L.L.P, Prabhu, Y, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Dev.Cell, 18, 2010
|
|
2G3Q
 
 | |
3LIQ
 
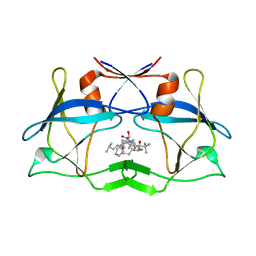 | | Crystal Structure of HTLV protease complexed with the inhibitor, KNI-10673 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-(2-methylpropyl)-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
2G6B
 
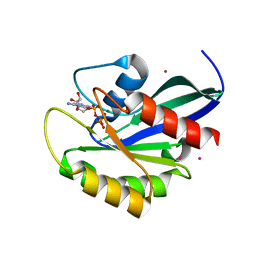 | | Crystal structure of human RAB26 in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-26, ... | | Authors: | Wang, J, Tempel, W, Shen, Y, Shen, L, Yaniw, D, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-24 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human RAB26 in complex with a GTP analogue
To be Published
|
|
3LJO
 
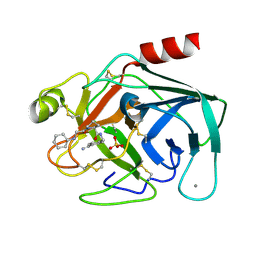 | | Bovine trypsin in complex with UB-THR 11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Wegscheid-Gerlach, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties.
J.Mol.Biol., 405, 2011
|
|
3LIY
 
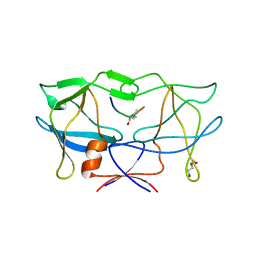 | | crystal structure of HTLV protease complexed with Statine-containing peptide inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease, statine-containing inhibitor | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
4L17
 
 | | GluA2-L483Y-A665C ligand-binding domain in complex with the antagonist DNQX | | Descriptor: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | Authors: | Lau, A.Y, Blachowicz, L, Roux, B. | | Deposit date: | 2013-06-02 | | Release date: | 2013-08-14 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A conformational intermediate in glutamate receptor activation.
Neuron, 79, 2013
|
|
2G08
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product-transition complex analog with Aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
3LN2
 
 | |
7ZAY
 
 | | Human heparan sulfate polymerase complex EXT1-EXT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-1, Exostosin-2, ... | | Authors: | Leisico, F, Omeiri, J, Hons, M, Schoehn, G, Lortat-Jacob, H, Wild, R. | | Deposit date: | 2022-03-23 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human heparan sulfate polymerase complex EXT1-EXT2.
Nat Commun, 13, 2022
|
|
