8DKM
 
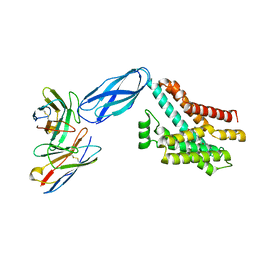 | |
8SDM
 
 | | HTRA-1 PDSA bound to CKP 3B3 | | Descriptor: | Cysteine-containing peptide 3B3, Serine protease HTRA1 | | Authors: | Ultsch, M.H, Kirchhofer, D, Wei, Y. | | Deposit date: | 2023-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Cystine-knot peptide inhibitors of HTRA1 bind to a cryptic pocket within the active site region.
Nat Commun, 15, 2024
|
|
5ZSS
 
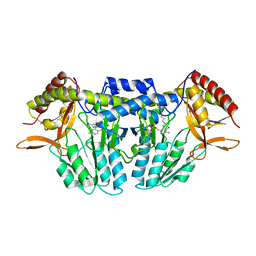 | |
8SE7
 
 | | HTRA-1 PDSA bound to CKP 1A8 | | Descriptor: | Cysteine knot peptide, Serine protease HTRA1 | | Authors: | Ultsch, M.H, Kirchhofer, D, Wei, Y. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Cystine-knot peptide inhibitors of HTRA1 bind to a cryptic pocket within the active site region.
Nat Commun, 15, 2024
|
|
8SE8
 
 | | HTRA-1 PD/SA bound to CKP 1G10 | | Descriptor: | Cysteine knot peptide, GLYCEROL, SULFATE ION, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cystine-knot peptide inhibitors of HTRA1 bind to a cryptic pocket within the active site region.
Nat Commun, 15, 2024
|
|
8SDP
 
 | | HTRA-1 PDSA bound to CKP 3A7 | | Descriptor: | Cysteine knot peptide 3A7, SULFATE ION, Serine protease HTRA1 | | Authors: | Ultsch, M.H, Kirchhofer, D, Wei, Y. | | Deposit date: | 2023-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Cystine-knot peptide inhibitors of HTRA1 bind to a cryptic pocket within the active site region.
Nat Commun, 15, 2024
|
|
1RN7
 
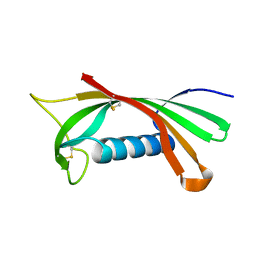 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1C7N
 
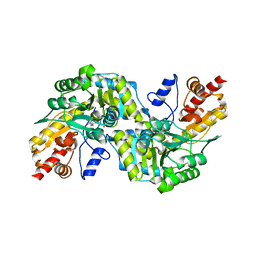 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE COFACTOR | | Descriptor: | CYSTALYSIN, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1ROA
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
2HM3
 
 | | Nematocyst outer wall antigen, cysteine rich domain NW1 | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Sequence-Structure and Structure-Function Analysis in Cysteine-rich Domains Forming the Ultrastable Nematocyst Wall.
J.Mol.Biol., 368, 2007
|
|
3NX0
 
 | | Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C | | Descriptor: | Cystatin-C, SULFATE ION | | Authors: | Orlikowska, M, Jankowska, E, Kolodziejczyk, R, Jaskolski, M, Szymanska, A. | | Deposit date: | 2010-07-12 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C.
J.Struct.Biol., 173, 2011
|
|
2HZ3
 
 | |
2HZ2
 
 | |
2HZ1
 
 | |
8QNO
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase treated at 368 K from Pyrococcus furiosus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Koeppl, L.H, Popadic, D, Andexer, J.N. | | Deposit date: | 2023-09-27 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
5M5K
 
 | | S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine and cordycepin | | Descriptor: | 3'-DEOXYADENOSINE, ACETATE ION, ADENOSINE, ... | | Authors: | Manszewski, T, Mueller-Dieckamann, J, Jaskolski, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
8COD
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Saleem-Batcha, R, Popadic, D, Koeppl, L.H, Andexer, J.N. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
6G7F
 
 | | Yeast 20S proteasome in complex with Cystargolide B | | Descriptor: | CHLORIDE ION, Cystargolide B- bound form, MAGNESIUM ION, ... | | Authors: | Groll, M, Tello-Aburto, R. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLL
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLR
 
 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9FCL
 
 | | CysG(N-16) in complex with SAM from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
9FCY
 
 | | CysG(N-16)-R21K mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | ADENINE, CALCIUM ION, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
