8F4K
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.16 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4I
 
 | | RT XFEL structure of Photosystem II 2000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
7T65
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8F4D
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the third illumination at 2.15 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8G01
 
 | | YES Complex - E. coli MraY, Protein E ID21, E. coli SlyD | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase SlyD, GPE, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Riera, N. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
7SUS
 
 | | Crystal structure of Apelin receptor in complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Han, G.W, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8G02
 
 | | YES Complex - E. coli MraY, Protein E PhiX174, E. coli SlyD | | Descriptor: | Lysis protein E, Peptidyl-prolyl cis-trans isomerase, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Li, Y.E. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
6LN2
 
 | | Crystal structure of full length human GLP1 receptor in complex with Fab fragment (Fab7F38) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab7F38_heavy chain, Fab7F38_light chain, ... | | Authors: | Wu, F, Yang, L, Hang, K, Laursen, M, Wu, L, Han, G.W, Ren, Q, Roed, N.K, Lin, G, Hanson, M, Jiang, H, Wang, M, Reedtz-Runge, S, Song, G, Stevens, R.C. | | Deposit date: | 2019-12-28 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full-length human GLP-1 receptor structure without orthosteric ligands.
Nat Commun, 11, 2020
|
|
8F4E
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the third illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8FW5
 
 | | Chimeric HsGATOR1-SpGtr-SpLam complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Tettoni, S.D, Egri, S.B, Doxsey, D.D, Ouch, C, Chang, J, Song, K, Xu, C, Shen, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the Schizosaccharomyces pombe Gtr-Lam complex reveals evolutionary divergence of mTORC1-dependent amino acid sensing.
Structure, 31, 2023
|
|
8F4G
 
 | | RT XFEL structure of Photosystem II 730 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8FVG
 
 | |
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8H07
 
 | |
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
8H9J
 
 | | Human ATP synthase state2 subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
6LQY
 
 | | Crystal complex of endo-deglycosylated hydroxynitrile lyase isozyme 5 of Prunus communis with benzaldehyde | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PREDICTED: (R)-mandelonitrile lyase, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-01-15 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
8ET5
 
 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase S653T mutant in complex with amidosulfuron | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of the Commercial Herbicide, Amidosulfuron, in Complex with Arabidopsis thaliana Acetohydroxyacid Synthase.
J.Agric.Food Chem., 71, 2023
|
|
8H9T
 
 | | Human ATP synthase state 2 (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
6LR8
 
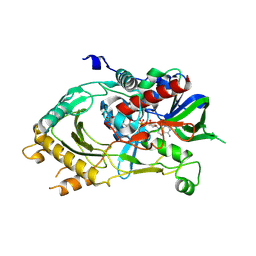 | |
8GZU
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory Megacomplex MC (IV2+I+III2+II)2 | | Descriptor: | 2 iron, 2 sulfur cluster-binding protein, 2-oxoglutarate/malate carrier protein, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8H2U
 
 | | X-ray Structure of photosystem I-LHCI super complex from Chlamydomonas reinhardtii. | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, H, Kubota-Kawai, H, Misumi, Y, Kurisu, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
8GYM
 
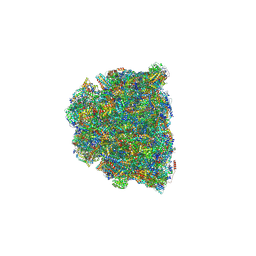 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8HL9
 
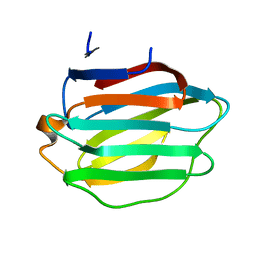 | | Crystal structure of Galectin-8 C-CRD with part of linker | | Descriptor: | Galectin-8 | | Authors: | Si, Y.L. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Linker remodels human Galectin-8 structure and regulates its hemagglutination and pro-apoptotic activity.
Int.J.Biol.Macromol., 245, 2023
|
|
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
