3ODS
 
 | |
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
3O51
 
 | | Crystal structure of anthranilamide 10 bound to AuroraA | | Descriptor: | N-[4-({3-[5-fluoro-2-(methylideneamino)pyrimidin-4-yl]pyridin-2-yl}oxy)phenyl]-2-(phenylamino)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
1U2V
 
 | | Crystal structure of Arp2/3 complex with bound ADP and calcium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-Related Protein 2, Actin-Related Protein 3, ... | | Authors: | Nolen, B.J, Littlefield, R.S, Pollard, T.D. | | Deposit date: | 2004-07-20 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1U7B
 
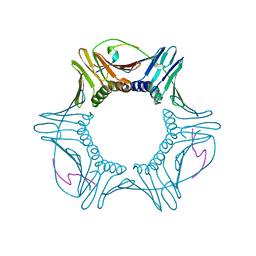 | |
3RQG
 
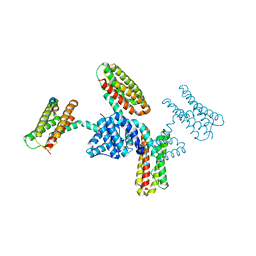 | |
3RI4
 
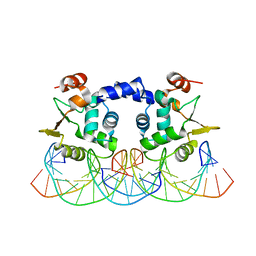 | |
3OB7
 
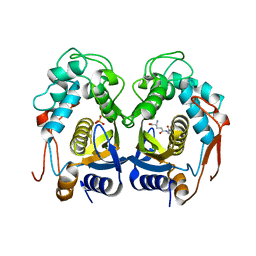 | | Human Thymidylate Synthase R163K with Cys 195 covalently modified by Glutathione | | Descriptor: | GLUTATHIONE, PHOSPHATE ION, Thymidylate synthase | | Authors: | Gibson, L.M, Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2010-08-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1TE6
 
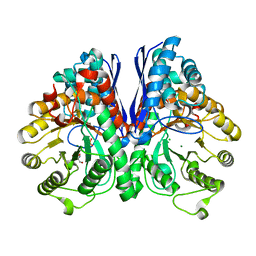 | | Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Gamma enolase, ... | | Authors: | Chai, G, Brewer, J, Lovelace, L, Aoki, T, Minor, W, Lebioda, L. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Purification and the 1.8 A Resolution Crystal Structure of Human Neuron Specific Enolase
J.Mol.Biol., 341, 2004
|
|
1TEV
 
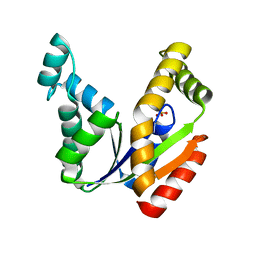 | | Crystal structure of the human UMP/CMP kinase in open conformation | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Segura-Pena, D, Sekulic, N, Ort, S, Konrad, M, Lavie, A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced Conformational Changes in Human UMP/CMP Kinase.
J.Biol.Chem., 279, 2004
|
|
3ODI
 
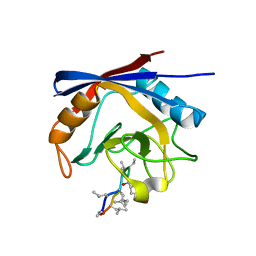 | | Crystal structure of cyclophilin A in complex with Voclosporin E-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ODL
 
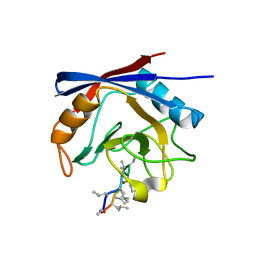 | | Crystal structure of cyclophilin A in complex with Voclosporin Z-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O8V
 
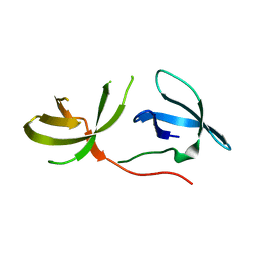 | | Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | Fragile X mental retardation syndrome-related protein 1 | | Authors: | Lam, R, Guo, Y.H, Adams-Cioaba, M, Bian, C.B, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
3R22
 
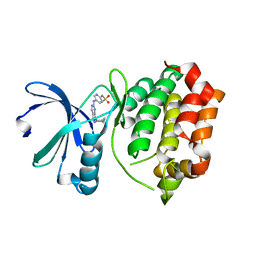 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RNU
 
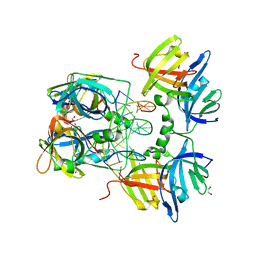 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
1U3J
 
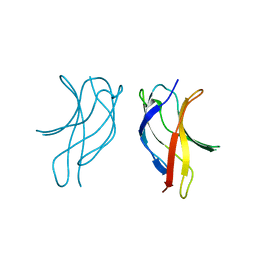 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
3ODR
 
 | |
3QBN
 
 | |
3Q6S
 
 | |
1TYQ
 
 | | Crystal structure of Arp2/3 complex with bound ATP and calcium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related Protein 2, Actin-related protein 3, ... | | Authors: | Nolen, B.J, Littlefield, R.S, Pollard, T.D. | | Deposit date: | 2004-07-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1UBD
 
 | | CO-CRYSTAL STRUCTURE OF HUMAN YY1 ZINC FINGER DOMAIN BOUND TO THE ADENO-ASSOCIATED VIRUS P5 INITIATOR ELEMENT | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*CP*TP*CP*CP*AP*TP*TP*TP*TP*GP*AP*A P*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*CP*AP*AP*AP*AP*TP*GP*GP*AP*GP*AP*C P*CP*CP*T)-3'), PROTEIN (YY1 ZINC FINGER DOMAIN), ... | | Authors: | Houbaviy, H.B, Usheva, A, Shenk, T, Burley, S.K. | | Deposit date: | 1996-10-04 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1U76
 
 | |
3QSV
 
 | |
3RCW
 
 | | Crystal Structure of the bromodomain of human BRD1 | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, ACETATE ION, ... | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Felletar, I, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
