3BEC
 
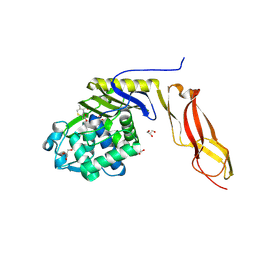 | |
3FSD
 
 | |
5URV
 
 | | Crystal structure of Frizzled 7 CRD in complex with C24 fatty acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
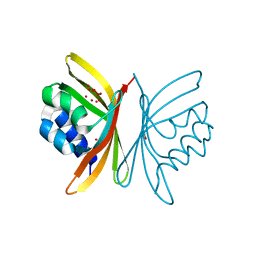
5URZ
 
 | | Crystal structure of Frizzled 5 CRD in complex with BOG | | Descriptor: | Frizzled-5, alpha-L-fucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, octyl beta-D-glucopyranoside | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
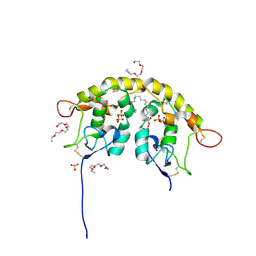
5URY
 
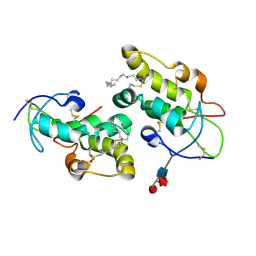 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | Descriptor: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W32
 
 | |
5W2Z
 
 | |
5W2K
 
 | |
5W2R
 
 | |
1EO8
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (HEAVY CHAIN), ... | | Authors: | Fleury, D, Gigant, B, Daniels, R.S, Skehel, J.J, Knossow, M, Bizebard, T. | | Deposit date: | 2000-03-22 | | Release date: | 2000-04-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for recognition of a single epitope by two distinct antibodies.
Proteins, 40, 2000
|
|
5W2X
 
 | |
5W31
 
 | |
5VNP
 
 | |
5C2X
 
 | | Crystal structure of deoxyribose-phosphate aldolase from Colwellia psychrerythraea (tetragonal form) | | Descriptor: | CARBONATE ION, Deoxyribose-phosphate aldolase, SULFATE ION, ... | | Authors: | Dick, M, Weiergraeber, O.H, Pietruszka, J. | | Deposit date: | 2015-06-16 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Trading off stability against activity in extremophilic aldolases.
Sci Rep, 6, 2016
|
|
3GI7
 
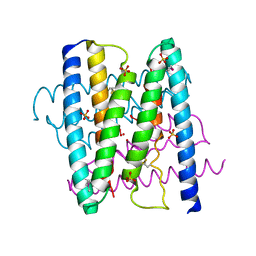 | |
3F7S
 
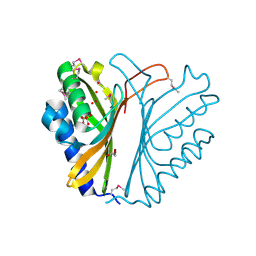 | |
3FF0
 
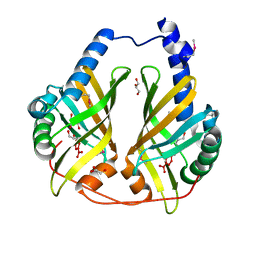 | |
3GE5
 
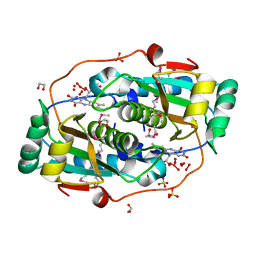 | |
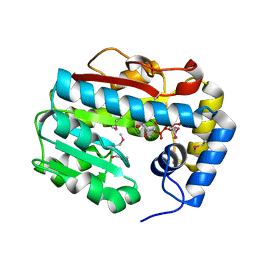
3G16
 
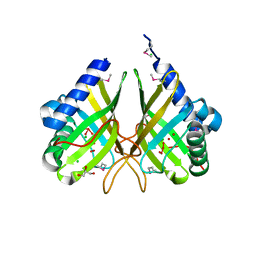 | |
3GBH
 
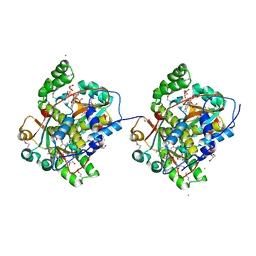 | |
3GIW
 
 | |
1WC2
 
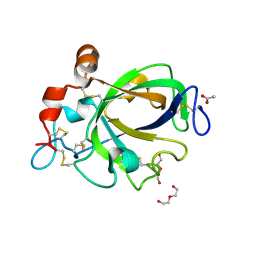 | | Beta-1,4-D-endoglucanase Cel45A from blue mussel Mytilus edulis at 1.2A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, ENDOGLUCANASE | | Authors: | Jakobsson, E, Mahdi, S, Kleywegt, G.J, Stahlberg, J. | | Deposit date: | 2004-11-08 | | Release date: | 2006-05-24 | | Last modified: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Glucomannan and beta-glucan degradation by Mytilus edulis Cel45A: Crystal structure and activity comparison with GH45 subfamily A, B and C.
Carbohydr Polym, 277, 2022
|
|
1NC2
 
 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA | | Descriptor: | (S)-2-(4-(2-(2-HYDROXYETHYLTHIO)-ACETAMIDO)-BENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
3EER
 
 | | High resolution structure of putative organic hydroperoxide resistance protein from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, IMIDAZOLE, Organic hydroperoxide resistance protein, ... | | Authors: | Nocek, B, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-05 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the organic hydroperoxide
resistance protein from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
2D1F
 
 | | Structure of Mycobacterium tuberculosis threonine synthase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine synthase | | Authors: | Covarrubias, A.S, Bergfors, T, Mannerstedt, K, Oscarson, S, Jones, T.A, Mowbray, S.L, Hogbom, M. | | Deposit date: | 2005-08-20 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical, and in vivo investigations of the threonine synthase from Mycobacterium tuberculosis.
J.Mol.Biol., 381, 2008
|
|
