6BUC
 
 | |
3LOY
 
 | | Crystal structure of a Copper-containing benzylamine oxidase from Hansenula Polymorpha | | Descriptor: | COPPER (II) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Klema, V.J, Johnson, B.J, Wilmot, C.M. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha.
Biochemistry, 49, 2010
|
|
1WYW
 
 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
7A6U
 
 | | Cryo-EM structure of the cytoplasmic domain of human TRPC6 | | Descriptor: | Short transient receptor potential channel 6, UNKNOWN ATOM OR ION | | Authors: | Grieben, M, Pike, A.C.W, Wang, D, Mukhopadhyay, S.M.M, Chalk, R, Marsden, B.D, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structure of the cytoplasmic domain of human TRPC6
TO BE PUBLISHED
|
|
6O5G
 
 | | Calmodulin in complex with isomalbrancheamide D | | Descriptor: | (5aS,12aS,13aS)-9-bromo-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7 ,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2019-03-02 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7PQE
 
 | |
7PPO
 
 | |
6OS4
 
 | | Calmodulin in complex with farnesyl cysteine methyl ester | | Descriptor: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | Authors: | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
2M55
 
 | | NMR structure of the complex of an N-terminally acetylated alpha-synuclein peptide with calmodulin | | Descriptor: | Alpha-synuclein, CALCIUM ION, Calmodulin | | Authors: | Gruschus, J.M, Yap, T, Pistolesi, S, Maltsev, A.S, Lee, J.C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Calmodulin Complexed to an N-Terminally Acetylated alpha-Synuclein Peptide.
Biochemistry, 52, 2013
|
|
1A5R
 
 | | STRUCTURE DETERMINATION OF THE SMALL UBIQUITIN-RELATED MODIFIER SUMO-1, NMR, 10 STRUCTURES | | Descriptor: | SUMO-1 | | Authors: | Bayer, P, Arndt, A, Metzger, S, Mahajan, R, Melchior, F, Jaenicke, R, Becker, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the small ubiquitin-related modifier SUMO-1.
J.Mol.Biol., 280, 1998
|
|
6YNU
 
 | | CaM-P458 complex (crystal form 1) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6YNS
 
 | | CaM-P458 complex (crystal form 2) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
1BDW
 
 | |
6YA9
 
 | | Crystal structure of rsGCaMP in the ON state (non-illuminated) | | Descriptor: | CALCIUM ION, rsCGaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-03-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
8AHS
 
 | |
1AV2
 
 | | Gramicidin A/CsCl complex, active as a dimer | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Burkhart, B.M, Li, N, Langs, D.A, Duax, W.L. | | Deposit date: | 1997-09-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Conducting Form of Gramicidin a is a Right-Handed Double-Stranded Double Helix.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6ZBI
 
 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
8B6Q
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 with an insertion of Calmodulin-M13 fusion at position 154-156 that mimic the structure of CaProLa, an calcium gated protein labeling technology | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase,Calmodulin-1,Haloalkane dehalogenase,Calmodulin-1,M13 peptide | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the haloalkane dehalogenase HaloTag7 with an insertion of Calmodulin-M13 fusion at position 154-156 that mimic the structure of CaProLa, an calcium gated protein labeling technology
To Be Published
|
|
8GM5
 
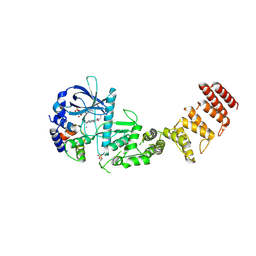 | | Functional construct of the Eukaryotic elongation factor 2 kinase bound to Calmodulin, ADP and to the A-484954 inhibitor and showing two conformations for the 498-520 loop | | Descriptor: | 7-amino-1-cyclopropyl-3-ethyl-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the complex between calmodulin and a functional construct of eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor.
J.Biol.Chem., 299, 2023
|
|
8GM4
 
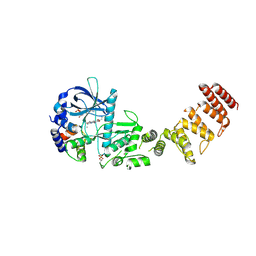 | | Functional construct of the Eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor | | Descriptor: | 7-amino-1-cyclopropyl-3-ethyl-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the complex between calmodulin and a functional construct of eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor.
J.Biol.Chem., 299, 2023
|
|
2ASQ
 
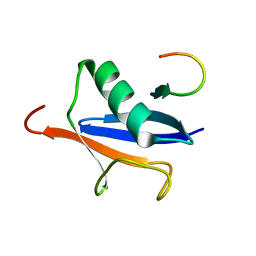 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
1QS0
 
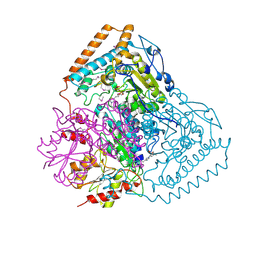 | | Crystal Structure of Pseudomonas Putida 2-oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B) | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-OXOISOVALERATE DEHYDROGENASE ALPHA-SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA-SUBUNIT, ... | | Authors: | Aevarsson, A, Seger, K, Turley, S, Sokatch, J.R, Hol, W.G.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-18 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes.
Nat.Struct.Biol., 6, 1999
|
|
6XXF
 
 | |
1Y8R
 
 | | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-SUMO1-MG-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-like 1 activating enzyme E1A, ... | | Authors: | Lois, L.M, Lima, C.D. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Embo J., 24, 2005
|
|
6MDO
 
 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
