6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8PMP
 
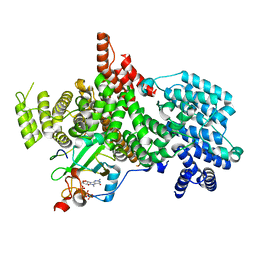 | | Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
6OWR
 
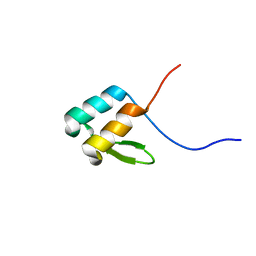 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
6OZ3
 
 | |
6YAM
 
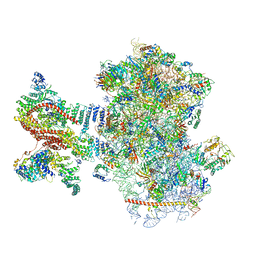 | | Mammalian 48S late-stage translation initiation complex (LS48S+eIF3 IC) with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6M4I
 
 | |
8TQD
 
 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
1C3E
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
6XAV
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
1C2T
 
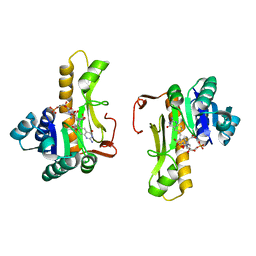 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
8R02
 
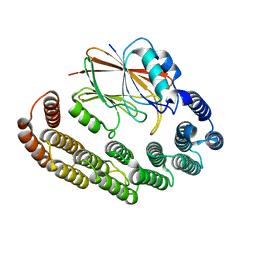 | | Crystal structure of the retromer complex VPS29/VPS35 with the ligand bis-1,3-phenyl guanylhydrazone, 2a | | Descriptor: | Bis-1,3-phenyl guanylhydrazon, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Milani, M, Fagnani, E. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Comput Struct Biotechnol J, 23, 2024
|
|
8R0J
 
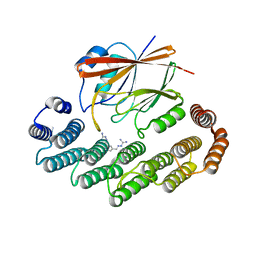 | | Crystal structure of the retromer complex VPS29/VPS35 with the ligand bis-1,3-phenyl guanylhydrazone, 2a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bis-1,3-phenyl guanylhydrazon, Vacuolar protein sorting-associated protein 29, ... | | Authors: | Milani, M, Fagnani, E. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Comput Struct Biotechnol J, 23, 2024
|
|
7NTJ
 
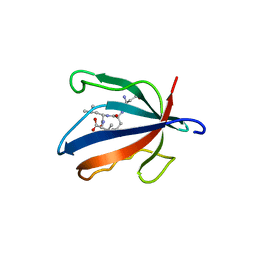 | | PALS1 PDZ1 domain with SARS-CoV-1_E PBM complex | | Descriptor: | Envelope small membrane protein, MAGUK p55 subfamily member 5 | | Authors: | Javorsky, A, Kvansakul, M. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain.
Commun Biol, 4, 2021
|
|
7O2V
 
 | | AURORA KINASE A IN COMPLEX WITH THE AUR-A/PDK1 INHIBITOR VI8 | | Descriptor: | 1-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N}-[(1~{R})-2-[[(3~{E})-3-(1~{H}-imidazol-5-ylmethylidene)-2-oxidanylidene-1~{H}-indol-5-yl]amino]-2-oxidanylidene-1-phenyl-ethyl]-6-methyl-2-oxidanylidene-pyridine-3-carboxamide, Aurora kinase A | | Authors: | Garau, G. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of potent dual PDK1/AurA kinase inhibitors for cancer therapy: Lead-optimization, structural insights, and ADME-Tox profile.
Eur.J.Med.Chem., 226, 2021
|
|
4Y4M
 
 | | Thiazole synthase Thi4 from Methanocaldococcus jannaschii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Putative ribose 1,5-bisphosphate isomerase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R)-2,3,5-tris(oxidanyl)-4-oxidanylidene-pentyl] hydrogen phosphate | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
8QFY
 
 | |
6Q9C
 
 | | Crystal structure of Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF bound to NADH under anaerobic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9G
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129D and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9J
 
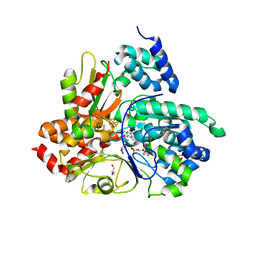 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129S and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6X4Y
 
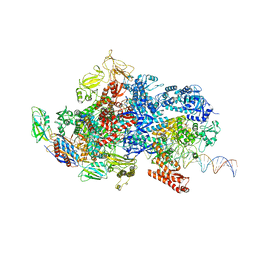 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
8A9Z
 
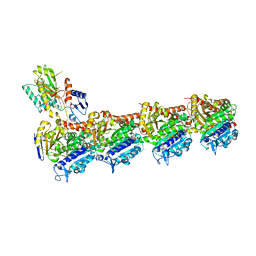 | | Tubulin-[1,2]oxazoloisoindole-2e complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-[(3,5-dimethoxyphenyl)methyl]pyrrolo[3,4-g][1,2]benzoxazole, CALCIUM ION, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
5HL4
 
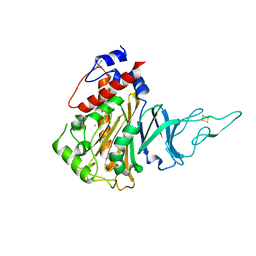 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
6X4W
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - III state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
5HQD
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
