1U3Y
 
 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
6MV8
 
 | | LDHA structure in complex with inhibitor 14 | | Descriptor: | (6S)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(2-hydroxyphenyl)-6-phenyl-5,6-dihydro-2H-pyran-2-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C.E, Ultsch, M, Wei, B. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based Optimization of Potent, Cell-Active
Hydroxylactam Inhibitors of Lactate Dehydrogenase
To Be Published
|
|
2XAS
 
 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2580, 14e) | | Descriptor: | 3-[4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)PHENYL]PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
6MH1
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH HU-10, A 1,4,5-Trisubstituted Imidazole Analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-(3,5-dimethylphenyl)-4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]pyrimidin-2-amine | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2018-09-17 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for the N-Terminal Bromodomain-and-Extra-Terminal-Family Selectivity of a Dual Kinase-Bromodomain Inhibitor.
J.Med.Chem., 61, 2018
|
|
2XA6
 
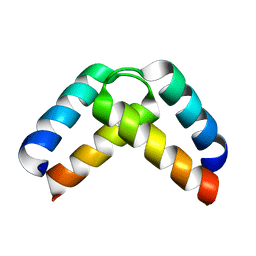 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING,RNA-BINDING,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
1R7R
 
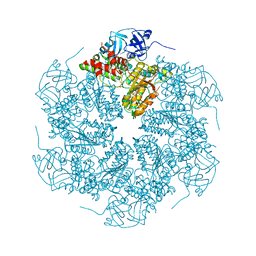 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
6MXY
 
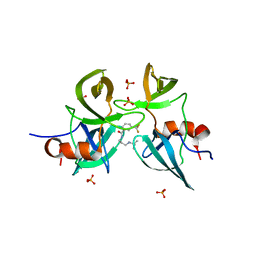 | |
1RTS
 
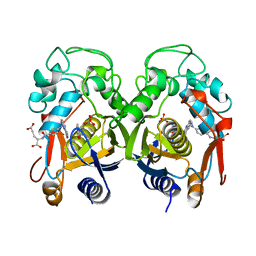 | | THYMIDYLATE SYNTHASE FROM RAT IN TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Sotelo-Mundo, R.R, Ciesla, J, Dzik, J.M, Rode, W, Maley, F, Maley, G, Hardy, L.W, Montfort, W.R. | | Deposit date: | 1998-06-19 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug.
Biochemistry, 38, 1999
|
|
6MRC
 
 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
1SJR
 
 | | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
6MXZ
 
 | | Structure of 53BP1 Tudor domains in complex with small molecule UNC3474 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-(propan-2-yl)benzamide, TP53-binding protein 1 | | Authors: | Cui, G, Botuyan, M.V, Schuller, D.J, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
2V5Q
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE PLK-1 KINASE DOMAIN IN COMPLEX WITH A SELECTIVE DARPIN | | Descriptor: | DESIGN ANKYRIN REPEAT PROTEIN, SERINE/THREONINE-PROTEIN KINASE PLK1 | | Authors: | Bandeiras, T.M, Hillig, R.C, Matias, P.M, Eberspaecher, U, Fanghaenel, J, Thomaz, M, Miranda, S, Crusius, K, Puetter, V, Amstutz, P, Gulotti-Georgieva, M, Binz, H.K, Holz, C, Schmitz, A.A.P, Lang, C, Donner, P, Egner, U, Carrondo, M.A, Mueller-Tiemann, B. | | Deposit date: | 2007-07-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of wild-type Plk-1 kinase domain in complex with a selective DARPin.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
2V1D
 
 | | Structural basis of LSD1-CoREST selectivity in histone H3 recognition | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3.1T, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Forneris, F, Binda, C, Adamo, A, Battaglioli, E, Mattevi, A. | | Deposit date: | 2007-05-23 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Lsd1-Corest Selectivity in Histone H3 Recognition.
J.Biol.Chem., 282, 2007
|
|
2W1F
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | N-[3-(1H-BENZIMIDAZOL-2-YL)-1H-PYRAZOL-4-YL]BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
6MTD
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U2V
 
 | | Crystal structure of Arp2/3 complex with bound ADP and calcium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-Related Protein 2, Actin-Related Protein 3, ... | | Authors: | Nolen, B.J, Littlefield, R.S, Pollard, T.D. | | Deposit date: | 2004-07-20 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2W1C
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-{[2-(4-{[(4-FLUOROPHENYL)CARBONYL]AMINO}-1H-PYRAZOL-3-YL)-1H-BENZIMIDAZOL-6-YL]METHYL}MORPHOLIN-4-IUM, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
1U7B
 
 | |
2W1G
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 2-{4-[(CYCLOPROPYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-6-(MORPHOLIN-4-IUM-4-YLMETHYL)-1H-3,1-BENZIMIDAZOL-3-IUM, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
6MWQ
 
 | |
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6N3C
 
 | | SegB, conformation of TDP-43 low complexity domain segment A | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2X2C
 
 | | acetyl-CypA:cyclosporine complex | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
