4OIT
 
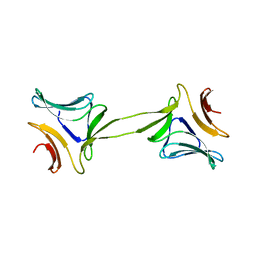 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
3UIT
 
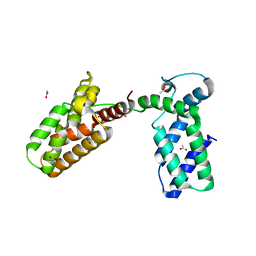 | | Overall structure of Patj/Pals1/Mals complex | | Descriptor: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | Authors: | Zhang, J, Yang, X, Long, J, Shen, Y. | | Deposit date: | 2011-11-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
4OU9
 
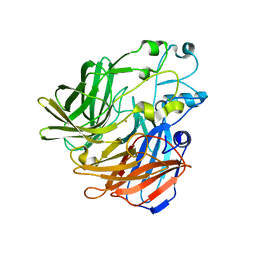 | | Crystal structure of apocarotenoid oxygenase in the presence of Triton X-100 | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Palczewski, K, Kiser, P.D. | | Deposit date: | 2014-02-15 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of Carotenoid Isomerase Activity in a Prototypical Carotenoid Cleavage Enzyme, Apocarotenoid Oxygenase (ACO).
J.Biol.Chem., 289, 2014
|
|
3UJM
 
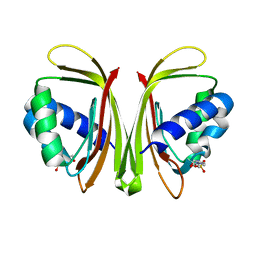 | |
4OJ5
 
 | |
3UJB
 
 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAH and phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphoethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJQ
 
 | | Galactose-specific lectin from Dolichos lablab in complex with galactose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shetty, K.N, Latha, V.L, Rao, R.N, Nadimpalli, S.K, Suguna, K. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Affinity of a galactose-specific legume lectin from Dolichos lablab to adenine revealed by X-ray cystallography.
Iubmb Life, 65, 2013
|
|
4OVU
 
 | | Crystal Structure of p110alpha in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
3UJO
 
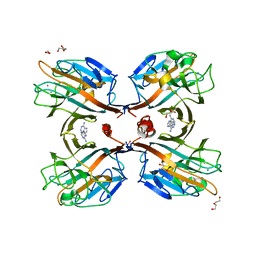 | | Galactose-specific seed lectin from Dolichos lablab in complex with adenine and galactose | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Shetty, K.N, Latha, V.L, Rao, R.N, Nadimpalli, S.K, Suguna, K. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity of a galactose-specific legume lectin from Dolichos lablab to adenine revealed by X-ray cystallography.
Iubmb Life, 65, 2013
|
|
4OLJ
 
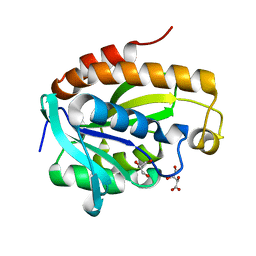 | | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA Hydrolase from Acinetobacter Baumannii at 1.49 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Sikarwar, J, Dube, D, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA hydrolase from Acinetobacter Baumannii at 1.49 A resolution
to be published
|
|
4OWW
 
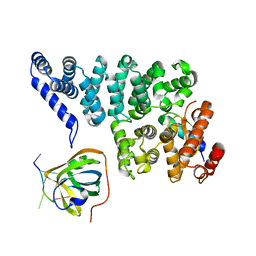 | | Structural basis of SOSS1 in complex with a 35nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1, ... | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
3UK4
 
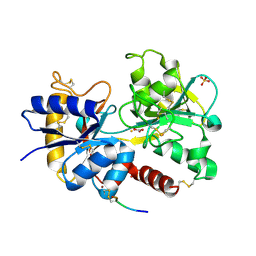 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with 1,2,5-Pentanetriol at 1.98 A Resolution | | Descriptor: | (2S)-pentane-1,2,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with 1,2,5-Pentanetriol at 1.98 A Resolution
To be Published
|
|
4OLU
 
 | | Crystal structure of antibody VRC07 in complex with clade A/E 93TH057 HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen binding fragment of heavy chain: Antibody VRC01, Antigen binding fragment of light chain: Antibody VRC01, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2014-01-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo.
J.Virol., 88, 2014
|
|
4OXQ
 
 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Zinc | | Descriptor: | Manganese ABC transporter, periplasmic-binding protein SitA, ZINC ION | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
3UKY
 
 | | Mouse importin alpha: yeast CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein complex subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UL7
 
 | | Crystal structure of the TV3 mutant F63W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
4OMC
 
 | |
4OO5
 
 | |
3ULS
 
 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULZ
 
 | | Crystal structure of apo BAK1 | | Descriptor: | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | | Authors: | Lou, Z.Y, Yan, L.M, Ma, Y.Y. | | Deposit date: | 2011-11-11 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for BAK1 activation
To be Published
|
|
4P0H
 
 | |
3ULF
 
 | |
4O6G
 
 | | Rv3902c from M. tuberculosis | | Descriptor: | Uncharacterized protein | | Authors: | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4OOA
 
 | | CRYSTAL STRUCTURE of NAF1 (MINER1): H114C THE REDOX-ACTIVE 2FE-2S PROTEIN | | Descriptor: | CDGSH iron-sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Tamir, S, Eisenberg-Domovich, Y, Conlan, A.R, Stofleth, J.T, Lipper, C.H, Paddock, M.L, Mittler, R, Jennings, P.A, Livnah, O, Nechushtai, R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3UMD
 
 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 4. | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
