7NHZ
 
 | |
6SX6
 
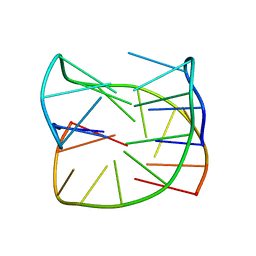 | | Guanine-rich oligonucleotide with 5'-GC end form G-quadruplex with A(GGGG)A hexad, GCGC- and G-quartets and two symmetric GG and AA base pairs | | Descriptor: | GCn | | Authors: | Pavc, D, Wang, B, Spindler, L, Drevensek-Olenik, I, Plavec, J, Sket, P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly.
Nucleic Acids Res., 48, 2020
|
|
7WKR
 
 | |
6SYK
 
 | | Guanine-rich oligonucleotide with 5'- and 3'-GC ends form G-quadruplex with A(GGGG)A hexad, GCGC- and G-quartets and two symmetric GG and AA base pair | | Descriptor: | GCnCG | | Authors: | Pavc, D, Wang, B, Spindler, L, Drevensek-Olenik, I, Plavec, J, Sket, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly.
Nucleic Acids Res., 48, 2020
|
|
8FZW
 
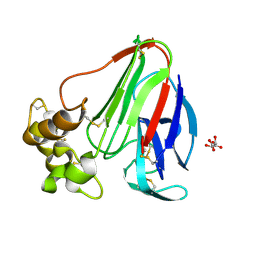 | | Thaumatin crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-01-30 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
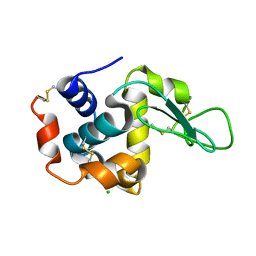
7NRN
 
 | | NMR structure of GIPC1-GH2 domain | | Descriptor: | PDZ domain-containing protein GIPC1 | | Authors: | Barthe, P, Roumestand, C. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pressure and Chemical Unfolding of an alpha-Helical Bundle Protein: The GH2 Domain of the Protein Adaptor GIPC1.
Int J Mol Sci, 22, 2021
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
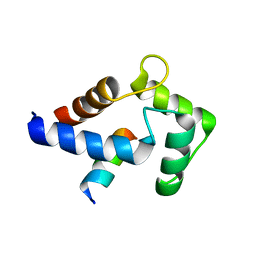
8DGK
 
 | |
7VRL
 
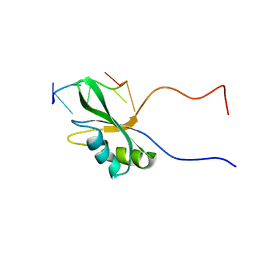 | | Solution structure of Rbfox RRM bound to a non-cognate RNA | | Descriptor: | RNA (5'-R(*UP*GP*CP*AP*UP*AP*U)-3'), RNA binding protein fox-1 homolog 1 | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2021-10-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity.
Nat Commun, 14, 2023
|
|
7WUC
 
 | |
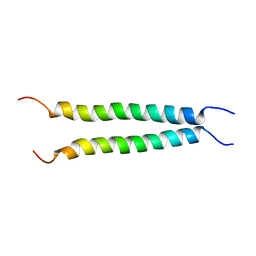
7VU5
 
 | |
6DMZ
 
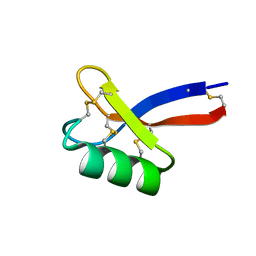 | | Solution structure of ZmD32 | | Descriptor: | Flower-specific gamma-thionin | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2018-06-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Salt-Tolerant Antifungal and Antibacterial Activities of the Corn Defensin ZmD32.
Front Microbiol, 10, 2019
|
|
6KJX
 
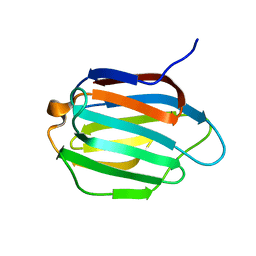 | | Galectin-13 variant C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
8U1U
 
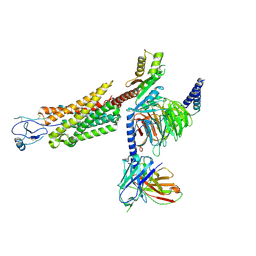 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6KHV
 
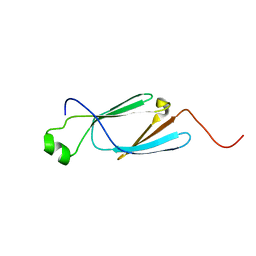 | | Solution Structure of the CS2 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
6JXV
 
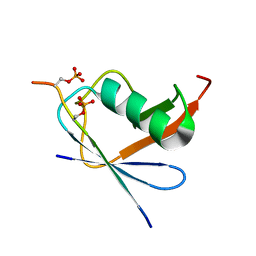 | | SUMO1 bound to phosphorylated SLS4-SIM peptide from ICP0 | | Descriptor: | Phosphorylated SLS4-SIM from ubiquitin E3 ligase ICP0, Small ubiquitin-related modifier | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6K84
 
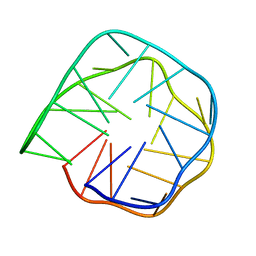 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
6JXX
 
 | | SUMO2 bound to phosphorylated SLS4-SIM peptide from ICP0 | | Descriptor: | Phosphorylated SLS4 from E3 ubiquitin ligase ICP0, Small ubiquitin-related modifier 2 | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6DT0
 
 | | Cryo-EM structure of a mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Yoo, J, Wu, M, Yin, Y, Herzik, M.A.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a mitochondrial calcium uniporter.
Science, 361, 2018
|
|
6D5Z
 
 | |
6D7T
 
 | | Cryo-EM structure of human TRPV6-Y467A in complex with 2-Aminoethoxydiphenyl borate (2-APB) | | Descriptor: | 2-aminoethyl diphenylborinate, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
8U4N
 
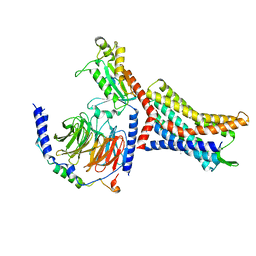 | | Structure of Apo CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
6KPF
 
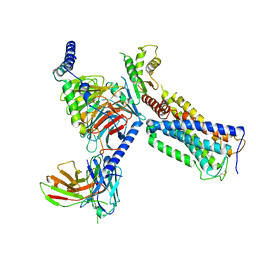 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
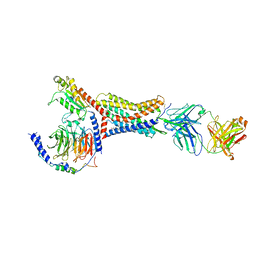
8U4Q
 
 | |
