1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
6QXH
 
 | |
6QXG
 
 | |
2XBJ
 
 | | Crystal Structure of Chk2 in complex with an inhibitor | | Descriptor: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Anderson, V.E, Walton, M.I, Eve, P.D, Caldwell, J.J, Pearl, L.H, Oliver, A.W, Collins, I, Garrett, M.D. | | Deposit date: | 2010-04-12 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
6R2E
 
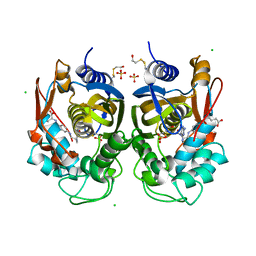 | | Crystal structure of the human thymidylate synthase (hTS) interface variant Q62R | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evidence of Destabilization of the Human Thymidylate Synthase (hTS) Dimeric Structure Induced by the Interface Mutation Q62R.
Biomolecules, 9, 2019
|
|
6M0W
 
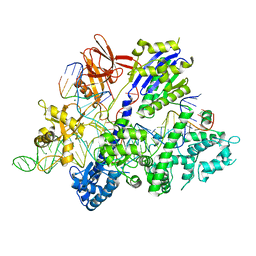 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | Descriptor: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
5ERC
 
 | |
6MFQ
 
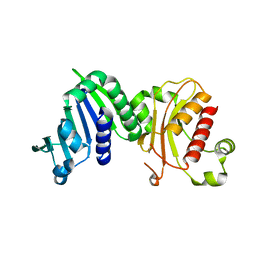 | | Crystal structure of a PMS2 variant | | Descriptor: | Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of two variants of uncertain significance in the PMS2 gene.
Hum. Mutat., 40, 2019
|
|
6M0V
 
 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-22 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
3MI7
 
 | | An Enhanced Repressor of Human Papillomavirus E2 Protein | | Descriptor: | 1,2-ETHANEDIOL, Regulatory protein E2 | | Authors: | Bohm, A, Baleja, J, Bose, K, Meinke, G. | | Deposit date: | 2010-04-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and characterization of an enhanced repressor of human papillomavirus E2 protein.
Faseb J., 25, 2011
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1OCP
 
 | | SOLUTION STRUCTURE OF OCT3 POU-HOMEODOMAIN | | Descriptor: | OCT-3 | | Authors: | Morita, E.H, Hayashi, F, Shirakawa, M, Kyogoku, Y. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Oct-3 POU-Homeodomain in Solution, as Determined by Triple Resonance Heteronuclear Multidimensional NMR Spectroscopy
Protein Sci., 4, 1995
|
|
3MXN
 
 | | Crystal structure of the RMI core complex | | Descriptor: | BENZAMIDINE, RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Hoadley, K.A, Xu, D, Xue, Y, Satyshur, K.A, Wang, W, Keck, J.L. | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome.
Structure, 18, 2010
|
|
6UD7
 
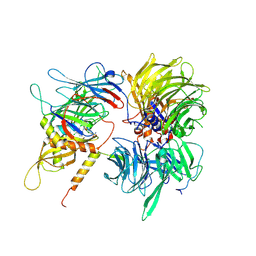 | | Crystal structure of full-length human DCAF15-DDB1(deltaBPB)-DDA1-RBM39 in complex with indisulam | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1,DNA damage-binding protein 1, ... | | Authors: | Bussiere, D.E, Shu, W, Xie, L, Knapp, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
1B4A
 
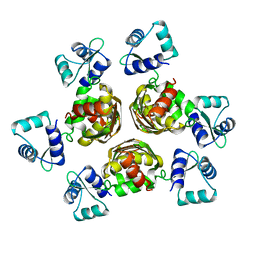 | | STRUCTURE OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
3IWZ
 
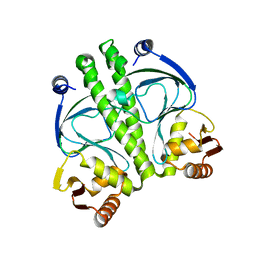 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
2CUJ
 
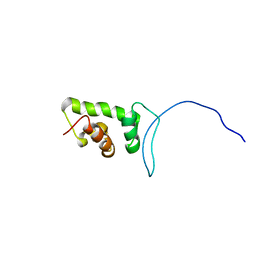 | | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like | | Descriptor: | transcriptional adaptor 2-like | | Authors: | Yoneyama, M, Umehara, T, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
6O5C
 
 | | X-ray crystal structure of metal-dependent transcriptional regulator MtsR | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Putative metal-dependent transcriptional regulator | | Authors: | Do, H, Kumaraswami, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Metal sensing and regulation of adaptive responses to manganese limitation by MtsR is critical for group A streptococcus virulence.
Nucleic Acids Res., 47, 2019
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5A4O
 
 | |
4AUX
 
 | | Tet repressor class D in complex with 9-nitrotetracycline | | Descriptor: | (4S,4aS,5aR,12aS)-4-(dimethylamino)-3,10,12,12a-tetrahydroxy-9-nitro-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Orth, P, Saenger, W, Hinrichs, W. | | Deposit date: | 2012-05-22 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tet Repressor Class D in Complex with 9-Nitrotetracycline and Magnesium
To be Published
|
|
6SC2
 
 | | Structure of the dynein-2 complex; IFT-train bound model | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3O74
 
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
5TZF
 
 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2 intermediate, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
