5K2F
 
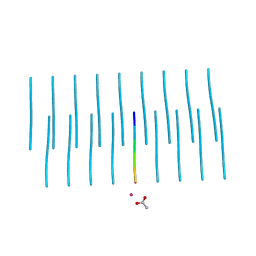 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2R
 
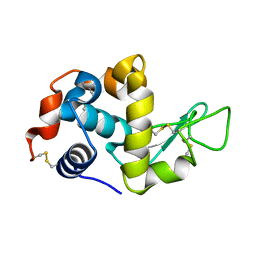 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of lysozyme with nano particles
To Be Published
|
|
6JCB
 
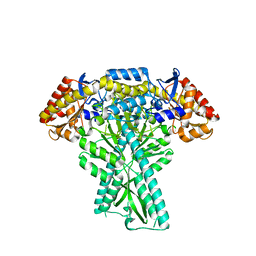 | |
5MY0
 
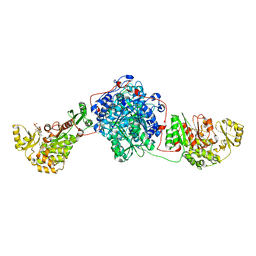 | | KS-MAT DI-DOMAIN OF MOUSE FAS WITH MALONYL-COA | | Descriptor: | COENZYME A, Fatty acid synthase, MALONYL-COENZYME A | | Authors: | Paithankar, K.S, Rittner, A, Huu, K.V, Grininger, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Characterization of the Polyspecific Transferase of Murine Type I Fatty Acid Synthase (FAS) and Implications for Polyketide Synthase (PKS) Engineering.
ACS Chem. Biol., 13, 2018
|
|
5MPS
 
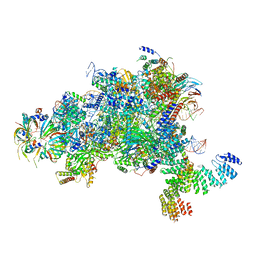 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
6JHJ
 
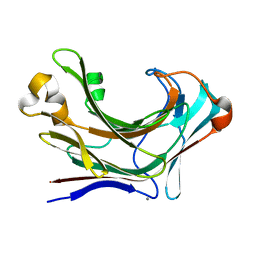 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
5MNV
 
 | |
5TUS
 
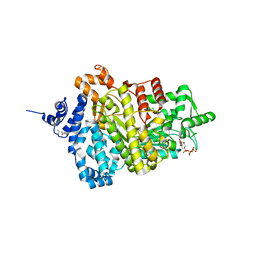 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5K1C
 
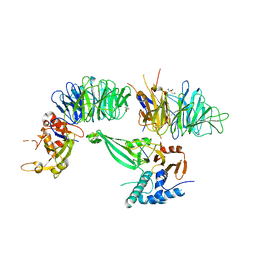 | | Crystal structure of the UAF1/WDR20/USP12 complex | | Descriptor: | PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Li, H, D'Andrea, A.D, Zheng, N. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allosteric Activation of Ubiquitin-Specific Proteases by beta-Propeller Proteins UAF1 and WDR20.
Mol.Cell, 63, 2016
|
|
1F5C
 
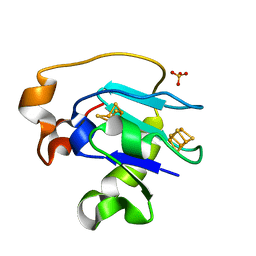 | | CRYSTAL STRUCTURE OF F25H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER, ... | | Authors: | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | Deposit date: | 2000-06-13 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5TQU
 
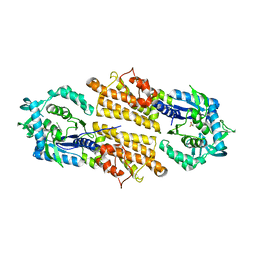 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor | | Descriptor: | 2-[2-[[3-(1~{H}-benzimidazol-2-ylamino)propylamino]methyl]-4,6-bis(chloranyl)indol-1-yl]ethanol, GLYCEROL, METHIONINE, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Cells to Mice to Target: Characterization of NEU-1053 (SB-443342) and Its Analogues for Treatment of Human African Trypanosomiasis.
ACS Infect Dis, 3, 2017
|
|
1F15
 
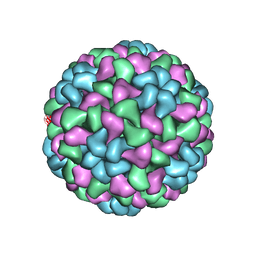 | |
5K2X
 
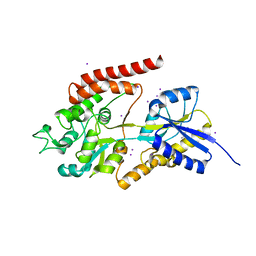 | | Crystal structure of M. tuberculosis UspC (tetragonal crystal form) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Futterer, K, Fullam, E, Besra, G.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional analysis of the solute-binding protein UspC from Mycobacterium tuberculosis that is specific for amino sugars.
Open Biology, 6, 2016
|
|
6J4D
 
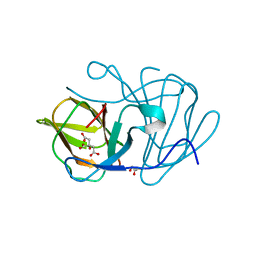 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under pH 4.7, without Zn | | Descriptor: | CITRATE ANION, Cupin superfamily protein, GLYCEROL | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for the Isomerization Mechanism of MarH
To Be Published
|
|
6JR7
 
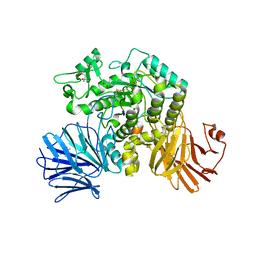 | | Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candidate alpha-glycosidase Glycoside hydrolase family 31, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2019-04-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into polysaccharide recognition by Flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31.
Febs J., 287, 2020
|
|
6JAE
 
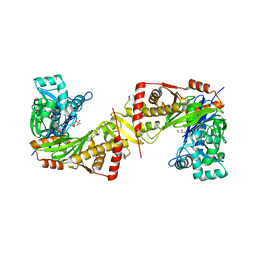 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with Pi (pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHATE ION | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
5TSO
 
 | |
5KBI
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH CATACHOL | | Descriptor: | CATECHOL, MopR, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
6JC5
 
 | |
1F5B
 
 | | CRYSTAL STRUCTURE OF F2H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | Deposit date: | 2000-06-13 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
5K7K
 
 | |
5MXJ
 
 | | Structure of the Y108F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
5MTW
 
 | |
6JF4
 
 | |
