4JKF
 
 | | Open and closed forms of T1791P+R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2GOM
 
 | |
4JKA
 
 | | Open and closed forms of R1865A human PRP8 RNase H-like domain with bound Co ion | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKG
 
 | | Open and closed forms of mixed T1789P+R1865A and R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KE8
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
5XVW
 
 | |
5XNG
 
 | | EFK17A structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-05-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5Y53
 
 | |
5XVJ
 
 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5XPI
 
 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
5YY9
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5YYA
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5XRX
 
 | | EFK17DA structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-06-10 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
5Y21
 
 | |
7E0N
 
 | | Crystal structure of Monoacylglycerol Lipase chimera | | Descriptor: | GLYCEROL, Thermostable monoacylglycerol lipase,Lipase | | Authors: | Lan, D.M. | | Deposit date: | 2021-01-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Monoacylglycerol Lipase chimera
To Be Published
|
|
7EBO
 
 | |
3ENB
 
 | |
3E9L
 
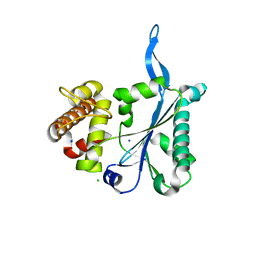 | | Crystal Structure of Human Prp8, Residues 1755-2016 | | Descriptor: | CHLORIDE ION, Pre-mRNA-processing-splicing factor 8, SODIUM ION | | Authors: | Pena, V, Rozov, A, Wahl, M.C. | | Deposit date: | 2008-08-22 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of an RNase H domain at the heart of the spliceosome.
Embo J., 27, 2008
|
|
8IK4
 
 | |
8IJP
 
 | |
3FNB
 
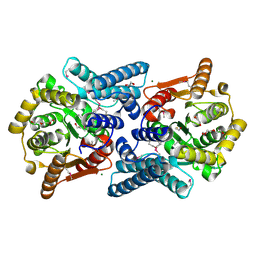 | | Crystal structure of acylaminoacyl peptidase SMU_737 from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, Acylaminoacyl peptidase SMU_737, BETA-MERCAPTOETHANOL, ... | | Authors: | Kim, Y, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-23 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1178 Å) | | Cite: | Crystal structure of acylaminoacyl-peptidase SMU_737 from Streptococcus mutans UA159
To be Published
|
|
8IKD
 
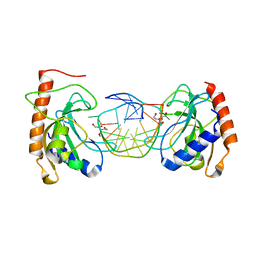 | |
8IK8
 
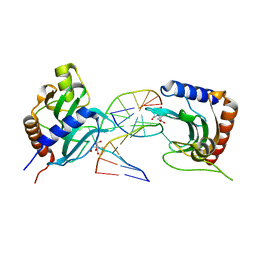 | |
8ILT
 
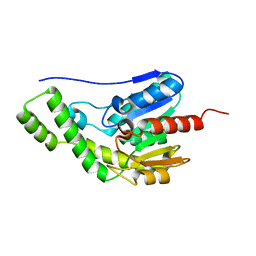 | | Crystal structure of Est30 | | Descriptor: | Carboxylesterase | | Authors: | Feng, Y, Luo, Z. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of Est30
To Be Published
|
|
