4K0C
 
 | | Crystal Structure of the computationally designed serine hydrolase. Northeast Structural Genomics Consortium (NESG) Target OR317 | | Descriptor: | designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR317
To be Published
|
|
2M5Q
 
 | |
5HFY
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-2-Ala24, beta-3-Lys28, beta-3-Lys31, beta-3-Asn35 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
7EKD
 
 | | Crystal structure of gibberellin 3-oxidase 2 (GA3ox2) in rice | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOGLUTARIC ACID, Gibberellin 3-beta-dioxygenase 2, ... | | Authors: | Takehara, S, Kawai, K, Mikami, B, Ueguchi-Tanaka, M. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Evolutionary alterations in gene expression and enzymatic activities of gibberellin 3-oxidase 1 in Oryza.
Commun Biol, 5, 2022
|
|
2MLE
 
 | | NMR structure of the C-domain of troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Baryshnikova, O.K, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
4KI9
 
 | | Crystal structure of the catalytic domain of human DUSP12 at 2.0 A resolution | | Descriptor: | Dual specificity protein phosphatase 12, PHOSPHATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-05-02 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6RMN
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8UF9
 
 | | EcDsbA in complex with 2-benzyl-4-phenylthiazole-5-carboxylic acid | | Descriptor: | 2-benzyl-4-phenyl-1,3-thiazole-5-carboxylic acid, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Two-mode inhibition of DsbA: combination of two site-specific inhibitors enhances virulence inhibition in Salmonella enterica serovar Typhimurium
To Be Published
|
|
1TLH
 
 | | T4 AsiA bound to sigma70 region 4 | | Descriptor: | 10 kDa anti-sigma factor, RNA polymerase sigma factor rpoD | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Embo J., 23, 2004
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | Descriptor: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
2MLF
 
 | | NMR structure of the dilated cardiomyopathy mutation G159D in troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Baryshnikova, O.K, Robertson, I.M, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
5N2M
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
5HJN
 
 | |
2LUL
 
 | | Solution NMR Structure of PH Domain of Tyrosine-protein kinase Tec from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR3504C | | Descriptor: | Tyrosine-protein kinase Tec, ZINC ION | | Authors: | Liu, G, Xiao, R, Janjua, H, Hamilton, K, Shastry, R, Kohan, E, Acton, T.B, Everett, J.K, Lee, H, Pederson, K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PH Domain of Tyrosine-protein kinase Tec from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR3504C
To be Published
|
|
8F0L
 
 | |
8B8O
 
 | |
5NCL
 
 | | Crystal structure of the Cbk1-Mob2 kinase-coactivator complex with an SSD1 peptide | | Descriptor: | CBK1 kinase activator protein MOB2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein SSD1, ... | | Authors: | Gogl, G, Remenyi, A, Parker, B, Weiss, E. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Ndr/Lats Kinases Bind Specific Mob-Family Coactivators through a Conserved and Modular Interface.
Biochemistry, 2020
|
|
8ELA
 
 | | CTX-M-14 beta-lactamase mutant - N132A w MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lu, S, Palzkill, T, Hu, L, Prasad, B.V.V. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
3C6B
 
 | |
4KPJ
 
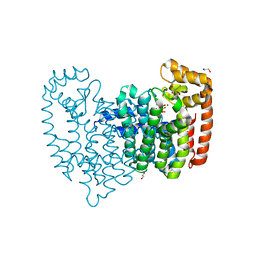 | | Crystal Structure of Farnesyl Pyrophosphate Synthase (Y204A) Mutant complexed with Mg, Pamidronate | | Descriptor: | 1,2-ETHANEDIOL, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C, Walter, R.L. | | Deposit date: | 2013-05-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Farnesyl Pyrophosphate Synthase (Y204A) Mutant complexed with Mg, Pamidronate
To be Published
|
|
6V1I
 
 | | Cryo-EM reconstruction of the thermophilic bacteriophage P74-26 small terminase- symmetric | | Descriptor: | Small terminase protein | | Authors: | Hayes, J.A, Hilbert, B.J, Gaubitz, C, Stone, N.P, Kelch, B.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A thermophilic phage uses a small terminase protein with a fixed helix-turn-helix geometry.
J.Biol.Chem., 295, 2020
|
|
7EMM
 
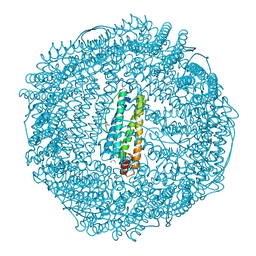 | | Crystal structure of IrCp* immobilized apo-R52H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Taher, M, Maity, B, Nakane, T, Abe, S, Ueno, T, Mazumdar, S. | | Deposit date: | 2021-04-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Controlled Uptake of an Iridium Complex inside Engineered apo-Ferritin Nanocages: Study of Structure and Catalysis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8OMZ
 
 | |
1TFE
 
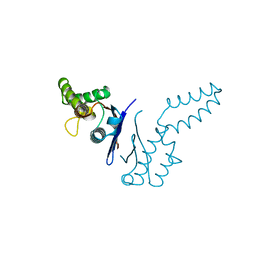 | | DIMERIZATION DOMAIN OF EF-TS FROM T. THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS | | Authors: | Jiang, Y, Nock, S, Nesper, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and importance of the dimerization domain in elongation factor Ts from Thermus thermophilus.
Biochemistry, 35, 1996
|
|
