3IMQ
 
 | | Crystal structure of the NusB101-S10(delta loop) complex | | Descriptor: | 30S ribosomal protein S10, N utilization substance protein B, POTASSIUM ION | | Authors: | Luo, X, Wahl, M.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination.
Nucleic Acids Res., 38, 2010
|
|
4IIN
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+ | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+
To be Published
|
|
4G8N
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Kastrup, J.S, Frydenvang, K, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
4KCT
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with Oxaloacetate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
4KCV
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with 2-oxoglutaric acid | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 2-OXOGLUTARIC ACID, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
4IP0
 
 | | X-Ray Structure of the Complex Uridine Phosphorylase from Vibrio cholerae with Phosphate Ion at 1.29 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-01-09 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
4KCW
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with oxalate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
4KRZ
 
 | | Apo crystal structure of pyruvate kinase (PYK) from Trypanosoma cruzi | | Descriptor: | GLYCEROL, POTASSIUM ION, Pyruvate kinase | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
4KS0
 
 | | Pyruvate kinase (PYK) from Trypanosoma cruzi in the presence of Magnesium, oxalate and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Morgan, H.P, Zhong, W, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
4KCU
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with D-Malate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, D-MALATE, MAGNESIUM ION, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
4HYV
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium, PEP and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HYW
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MH5
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
2GDI
 
 | |
4QK8
 
 | | Thermoanaerobacter pseudethanolicus c-di-AMP riboswitch | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, C-di-AMP riboswitch, MAGNESIUM ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into recognition of c-di-AMP by the ydaO riboswitch.
Nat.Chem.Biol., 10, 2014
|
|
2GRB
 
 | |
4QKA
 
 | | c-di-AMP riboswitch from Thermoanaerobacter pseudethanolicus, iridium hexamine soak | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, C-di-AMP riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into recognition of c-di-AMP by the ydaO riboswitch.
Nat.Chem.Biol., 10, 2014
|
|
3CCO
 
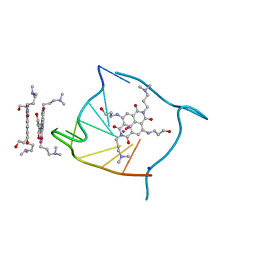 | | Structural adaptation and conservation in quadruplex-drug recognition | | Descriptor: | 2,7-bis[3-(dimethylamino)propyl]-4,9-bis[(3-hydroxypropyl)amino]benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, DNA (5'-D(*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), POTASSIUM ION, ... | | Authors: | Parkinson, G.N, Neidle, S. | | Deposit date: | 2008-02-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes
J.Mol.Biol., 381, 2008
|
|
3DCL
 
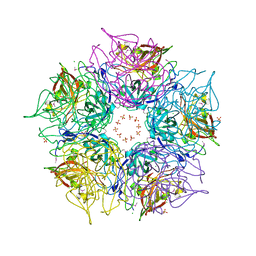 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
1E12
 
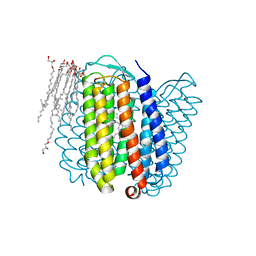 | | Halorhodopsin, a light-driven chloride pump | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, HALORHODOPSIN, ... | | Authors: | Essen, L.-O, Kolbe, M, Oesterhelt, D. | | Deposit date: | 2000-04-14 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Light-Driven Chloride Pump Halorhodopsin at 1.8 A Resolution
Science, 288, 2000
|
|
3DKE
 
 | | Polar and non-polar cavities in phage T4 lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AZIDE ION, ... | | Authors: | Liu, L.J, Matthews, B.W. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Use of experimental crystallographic phases to examine the hydration of polar and nonpolar cavities in T4 lysozyme
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | Descriptor: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | Authors: | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1N0X
 
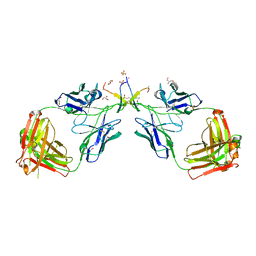 | | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, B2.1 peptide, GLYCEROL, ... | | Authors: | Saphire, E.O, Montero, M, Menendez, A, Irving, M.B, Zwick, M.B, Parren, P.W.H.I, Burton, D.R, Scott, J.K, Wilson, I.A. | | Deposit date: | 2002-10-15 | | Release date: | 2004-04-13 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope
To be Published
|
|
