6NLP
 
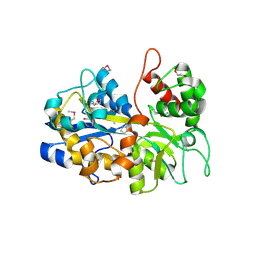 | | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113 | | Descriptor: | 1,2-ETHANEDIOL, Bacterial extracellular solute-binding family protein, IMIDAZOLE | | Authors: | Tan, K, SKarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113
To Be Published
|
|
8DFR
 
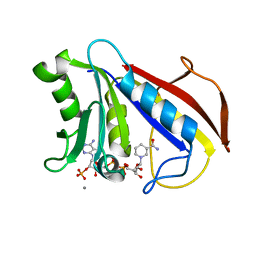 | | REFINED CRYSTAL STRUCTURES OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE. 3 ANGSTROMS APO-ENZYME AND 1.7 ANGSTROMS NADPH HOLO-ENZYME COMPLEX | | Descriptor: | CALCIUM ION, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Matthews, D.A, Oatley, S.J, Burridge, J, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1989-05-30 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined Crystal Structures of Chicken Liver Dihydrofolate Reductase. 3 Angstroms Apo-Enzyme and 1.7 Angstroms Nadph Holo-Enzyme Complex
To be Published
|
|
8ZCR
 
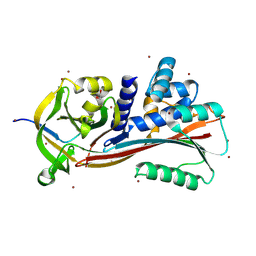 | | Structure of PI9 | | Descriptor: | 1,2-ETHANEDIOL, Serpin B9, ZINC ION | | Authors: | Zhou, A, Yan, T. | | Deposit date: | 2024-04-30 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization and crystallographic studies of human serine protease inhibitor (serpin) B9.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
4X01
 
 | | S. pombe Ctp1 tetramerization domain | | Descriptor: | 1,2-ETHANEDIOL, DNA binding ctp1 | | Authors: | Andres, S.N, Williams, R.S. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8U78
 
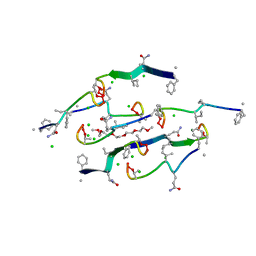 | | Structure of a N-Me-D-Gln4,Lys10-teixobactin analogue | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, N-Methyl-D-Gln4,Lys10-teixobactin | | Authors: | Nowick, J.S, Yang, H, Kreutzer, A.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
J.Org.Chem., 89, 2024
|
|
8DQ3
 
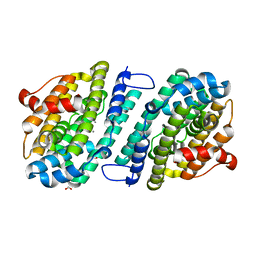 | |
4YJE
 
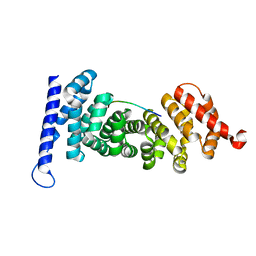 | | Crystal structure of APC-ARM in complexed with Amer1-A1 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
6BY5
 
 | | Two-State 14-mer UUCG Tetraloop calculated from Exact NOEs (State one: Conformers 1-5, State Two: Conformers 6-10) | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
9PFL
 
 | | Crystal structure of human 15-PGDH in complex with small molecule compound 1 | | Descriptor: | (4S)-7-[7-(4,4-difluoropiperidine-1-carbonyl)-2,3-dihydro-4H-1,4-benzoxazin-4-yl]-2-methyl[1,2,4]triazolo[4,3-a]pyridin-3(2H)-one, 1,2-ETHANEDIOL, 15-hydroxyprostaglandin dehydrogenase [NAD(+)], ... | | Authors: | Shaik, M.M, Brennan, D. | | Deposit date: | 2025-07-05 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Knowledge and Structure-Based Drug Design of 15-PGDH Inhibitors.
J.Med.Chem., 68, 2025
|
|
6Q1Y
 
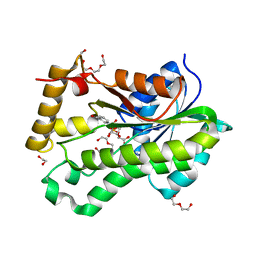 | |
5WV7
 
 | |
6NSY
 
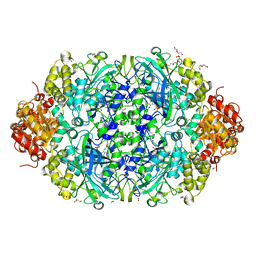 | | X-ray reduced Catalase 3 From N.Crassa in Cpd I state (0.263 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6NSZ
 
 | | X-ray reduced Catalase 3 from N.Crassa (0.526 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6NT1
 
 | | Catalase 3 from N.Crassa in ferrous state (2.89 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6O44
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-02-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
8DJM
 
 | | HMGCR-UBIAD1 Complex State 1 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
7RZP
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2866 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Dihydropteridine reductase, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
8DSG
 
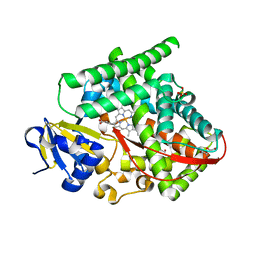 | | P411-PFA carbene transferase | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450-BM3 variant P411-PFA, ... | | Authors: | Maggiolo, A.O, Porter, N.J, Zhang, J, Arnold, F.H. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Chemodivergent C(sp 3 )-H and C(sp 2 )-H Cyanomethylation Using Engineered Carbene Transferases.
Nat Catal, 6, 2023
|
|
6O2X
 
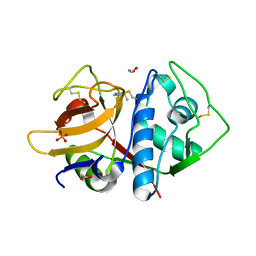 | | Structure of cruzain bound to MMTS inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6WZR
 
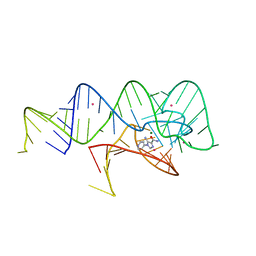 | | Fusibacterium ulcerans ZTP riboswitch bound to p-1-pyridinyl AICA | | Descriptor: | 5-amino-1-(pyridin-4-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | Authors: | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
6NSW
 
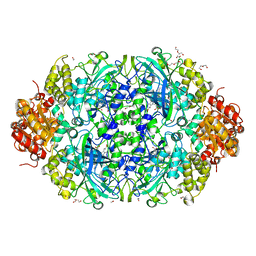 | | X-ray reduced Catalase 3 From N.Crassa in Cpd I state (0.135 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6OID
 
 | | Redox Regulation of FN3K from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Wood, Z.A, Kadirvelraj, R, Shrestha, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | A redox-active switch in fructosamine-3-kinases expands the regulatory repertoire of the protein kinase superfamily.
Sci.Signal., 13, 2020
|
|
8E8T
 
 | | Structure of the short LOR domain of human AASS | | Descriptor: | Alpha-aminoadipic semialdehyde synthase, mitochondrial | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2022-08-25 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Characterization and structure of the human lysine-2-oxoglutarate reductase domain, a novel therapeutic target for treatment of glutaric aciduria type 1.
Open Biology, 12, 2022
|
|
6YXD
 
 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YX9
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) at 2.4 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
