5DVX
 
 | | Crystal structure of the catalytic-domain of human carbonic anhydrase IX at 1.6 angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Carbonic anhydrase 9, ... | | Authors: | Mahon, B.P, Socorro, L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-09-21 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | The Structure of Carbonic Anhydrase IX Is Adapted for Low-pH Catalysis.
Biochemistry, 55, 2016
|
|
7VJP
 
 | | Selenomethionine-derived Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | SULFATE ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7Q16
 
 | | Human 14-3-3 zeta fused to the BAD peptide including phosphoserine-74 | | Descriptor: | 14-3-3 protein zeta/delta,Bcl2-associated agonist of cell death | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Gushchin, I, Remeeva, A, Kovalev, K, Cooley, R.B. | | Deposit date: | 2021-10-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Crystal structure of human 14-3-3 zeta complexed with the noncanonical phosphopeptide from proapoptotic BAD.
Biochem.Biophys.Res.Commun., 583, 2021
|
|
7PQ3
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in complex with its post-catalytic reaction product | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ2
 
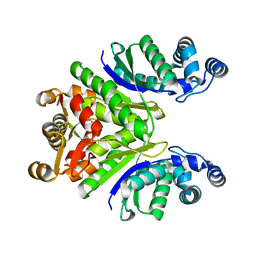 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
6DX2
 
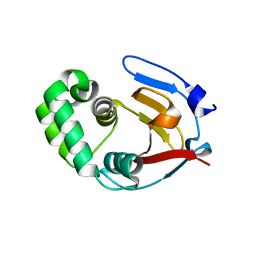 | | Crystal structure of the viral OTU domain protease from Dera Ghazi Khan virus | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Beldon, B.S, Dzimianski, J.V, Daczkowski, C.M, Goodwin, O.Y, Pegan, S.D. | | Deposit date: | 2018-06-28 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Probing the impact of nairovirus genomic diversity on viral ovarian tumor domain protease (vOTU) structure and deubiquitinase activity.
PLoS Pathog., 15, 2019
|
|
7PQ6
 
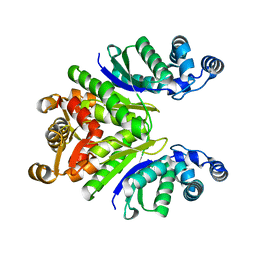 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12A from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQA
 
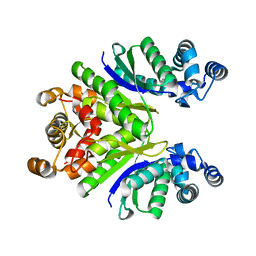 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12G/K169G from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
1DP0
 
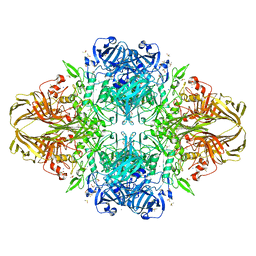 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
5KCG
 
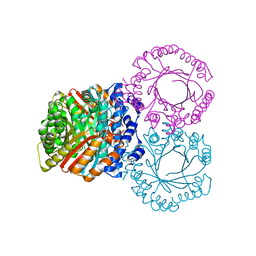 | |
5JWD
 
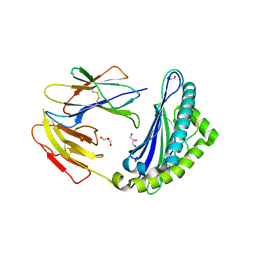 | | Crystal structure of H-2Db in complex with the LCMV-derived GP392-401 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
7Z5L
 
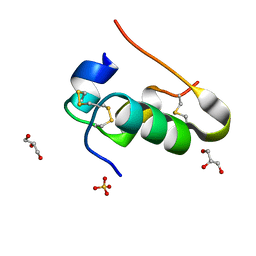 | |
8B37
 
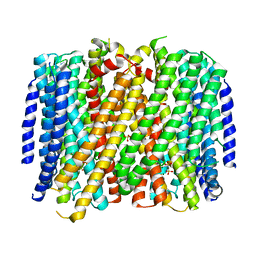 | | Crystal structure of Pyrobaculum aerophilum potassium-independent proton pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sulphate | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-insensitive pyrophosphate-energized proton pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Wilkinson, C, Vidilaseris, K, Ribeiro, O, Liu, J, Hillier, J, Malinen, A, Gehl, B, Jeuken, L.C, Pearson, A.R, Goldman, A. | | Deposit date: | 2022-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
7PGK
 
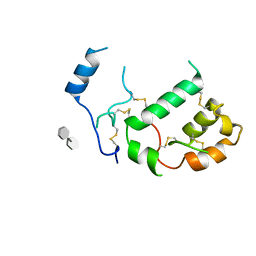 | | HHIP-N, the N-terminal domain of the Hedgehog-Interacting Protein (HHIP), in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
6IJI
 
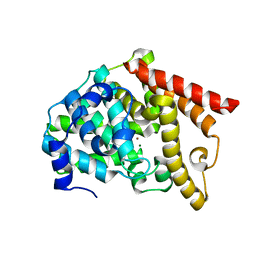 | | Crystal structure of PDE10 in complex with inhibitor 2b | | Descriptor: | 2-{2-[5-methyl-1-(pyridin-4-yl)-1H-benzimidazol-2-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
6A6J
 
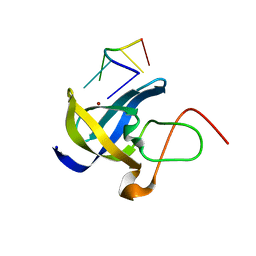 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
7ZBZ
 
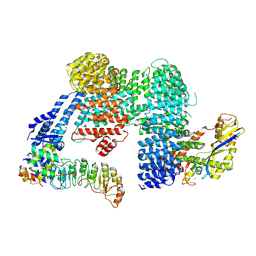 | |
6RFL
 
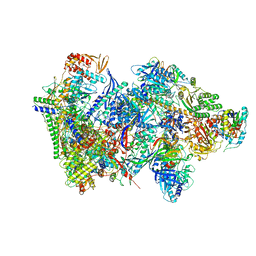 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
1O0V
 
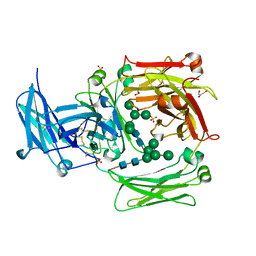 | | The crystal structure of IgE Fc reveals an asymmetrically bent conformation | | Descriptor: | GLYCEROL, Immunoglobulin heavy chain epsilon-1, SULFATE ION, ... | | Authors: | Wan, T, Beavil, R.L, Fabiane, S.M, Beavil, A.J, Sohi, M.K, Keown, M, Young, R.J, Henry, A.J, Owens, R.J, Gould, H.J, Sutton, B.J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of IgE Fc reveals an asymmetrically bent conformation
NAT.IMMUNOL., 3, 2002
|
|
6E06
 
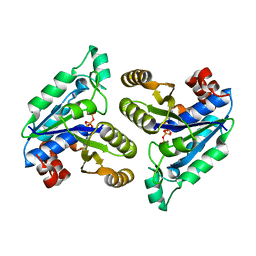 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in citrate precipitant) | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-07-06 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7Z8T
 
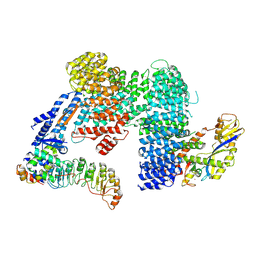 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8V
 
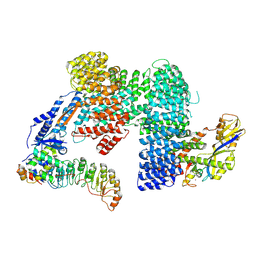 | | CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7ZBW
 
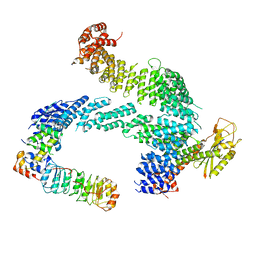 | | CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
6DZT
 
 | |
7DG5
 
 | |
