6ZQ4
 
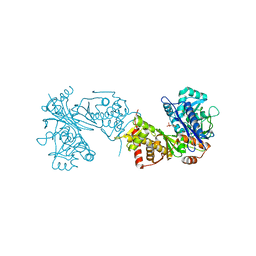 | |
3OXX
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, 1,2-ETHANEDIOL, ... | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3KMQ
 
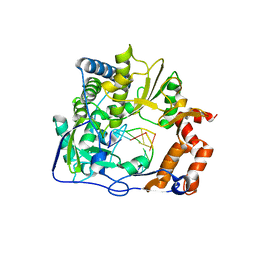 | | G62S mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA, tetragonal structure | | Descriptor: | 3D polymerase, RNA (5'-R(*GP*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*CP*C)-3') | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
5MYU
 
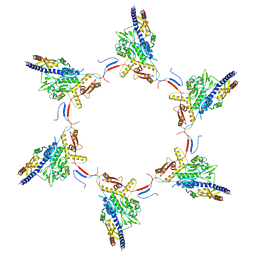 | | VipA-N2/VipB contracted sheath of type VI secretion system | | Descriptor: | Type VI secretion system protein ImpC, Uncharacterized protein | | Authors: | Wang, J, Brackmann, B, Castano-Diez, D, Kudryashev, M, Goldie, D, Maier, T, Stahlberg, H, Basler, M. | | Deposit date: | 2017-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Nat Microbiol, 2, 2017
|
|
1XEN
 
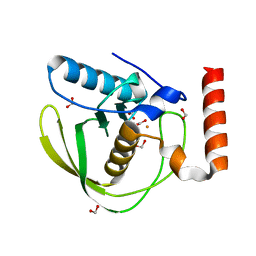 | | High Resolution Crystal Structure of Escherichia coli Iron- Peptide Deformylase Bound To Formate | | Descriptor: | FE (III) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
5N3F
 
 | |
1XNQ
 
 | |
1XI4
 
 | | Clathrin D6 Coat | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy
Nature, 432, 2004
|
|
3OX3
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(2-methoxy-1H-dipyrido[2,3-a:3',2'-e]pyrrolizin-11-yl)ethyl]furan-2-carboxamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
4QVN
 
 | | yCP beta5-M45V mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7BEL
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-88 and COVOX-45 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COVOX-45 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
4QWF
 
 | | yCP beta5-M45I mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3HLO
 
 | |
4P2O
 
 | | Crystal structure of the 2B4 TCR in complex with 2A/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2A peptide, 2B4 T-cell receptor alpha chain, ... | | Authors: | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
3HM3
 
 | |
4FJC
 
 | | Structure of the SAGA Ubp8/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | GLYCEROL, Protein SUS1, SAGA-associated factor 11, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
3PAW
 
 | |
1XY7
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G48480 | | Descriptor: | unknown protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-11-09 | | Release date: | 2004-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G48480
To be Published
|
|
3PAA
 
 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 8.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
1XMQ
 
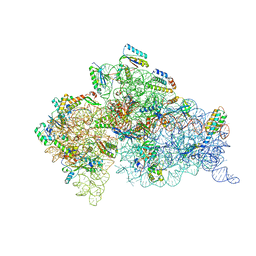 | | Crystal Structure of t6A37-ASLLysUUU AAA-mRNA Bound to the Decoding Center | | Descriptor: | 16s ribosomal RNA, 30S Ribosomal Protein S10, 30S Ribosomal Protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
4FQY
 
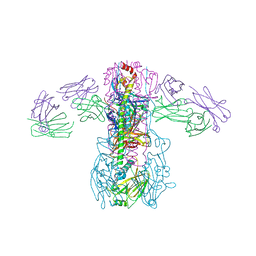 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H3 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.253 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FW2
 
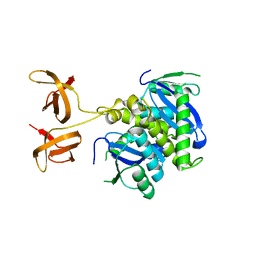 | |
3OKJ
 
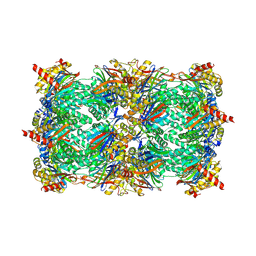 | | Alpha-keto-aldehyde binding mechanism reveals a novel lead structure motif for proteasome inhibition | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S,3S)-3-hydroxy-1-(4-hydroxyphenyl)-4-oxobutan-2-yl]-L-leucinamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Poynor, M, Gallastegui, P, Stein, M, Schmidt, B, Kloetzel, P.M, Huber, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elucidation of the alpha-keto-aldehyde binding mechanism: a lead structure motif for proteasome inhibition
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P0A
 
 | |
3I4U
 
 | |
