6GAY
 
 | |
8KAP
 
 | | Glycoside hydrolase family 1 beta-glucosidase from Streptomyces griseus (ligand-free) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Kumakura, H, Motouchi, S, Nakai, H, Nakajima, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of beta-1,2-glucan-associated glycoside hydrolase family 1 beta-glucosidase from Streptomyces griseus
To Be Published
|
|
6Y5F
 
 | | Crystal structure of the envelope glycoprotein prefusion complex of Andes virus - Mutant H953F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein,Envelope polyprotein, PHOSPHATE ION, ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2020-02-25 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism.
Cell, 183, 2020
|
|
6GB3
 
 | |
6UTA
 
 | | Crystal structure of Z004 iGL Fab in complex with ZIKV EDIII | | Descriptor: | Env, Z004 iGL Fab heavy chain, Z004 iGL Fab light chain | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6URV
 
 | | Crystal structure of Yellow Fever Virus NS2B-NS3 protease domain | | Descriptor: | NS2B, NS3 protease | | Authors: | Noske, G.D, Gawriljuk, V.F.O, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization and polymorphism analysis of the NS2B-NS3 protease from the 2017 Brazilian circulating strain of Yellow Fever virus.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6GB5
 
 | | Structure of H-2Db with truncated SEV peptide and GL | | Descriptor: | Beta-2-microglobulin, GLY-LEU, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P90
 
 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
8KDQ
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein -T03 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design protein -T03
To Be Published
|
|
6US9
 
 | |
6Y1L
 
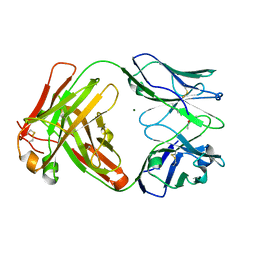 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P99
 
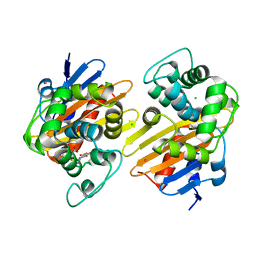 | | OXA-48 carbapanemase, ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
5HOX
 
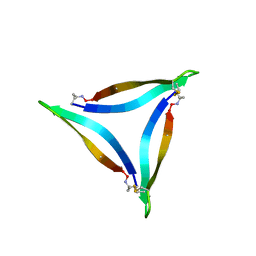 | | X-ray crystallographic structure of an A-beta 17_36 beta-hairpin. Synchrotron data set. (LVFFAEDCGSNKCAII(SAR)LMV). | | Descriptor: | Amyloid beta A4 protein, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Kreutzer, A.G, Nowick, J.S, Spencer, R.K. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an A beta 17-36 beta-Hairpin.
J.Am.Chem.Soc., 138, 2016
|
|
5D7I
 
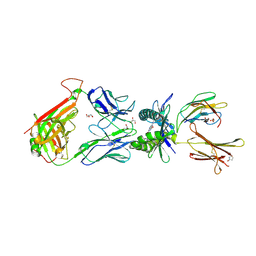 | | Structure of human MR1-Ac-6-FP in complex with human MAIT M33.64 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, M33.64 TCR Alpha Chain, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
5D8P
 
 | | 2.35A resolution structure of iron bound BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
7C5C
 
 | |
5DH2
 
 | |
5VJQ
 
 | | Complex between HyHEL10 Fab fragment heavy chain mutant (I29F, S52T, Y53F) and Pekin duck egg lysozyme isoform I (DEL-I) | | Descriptor: | CHLORIDE ION, GLYCEROL, HyHEL10 heavy chain Fab fragment carrying three mutations; I29F, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination.
Science, 360, 2018
|
|
8KBE
 
 | | Structure of CbTad1 complexed with 1',3'-cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Single phage proteins sequester signals from TIR and CGAS-like enzymes
Nature, 2024
|
|
6PAL
 
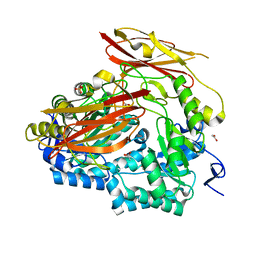 | | Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus | | Descriptor: | ACETATE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tamura, K, Brumer, H, van Petegem, F. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human GutBacteroides.
Mbio, 11, 2020
|
|
8KC1
 
 | | De novo design protein -NX5 | | Descriptor: | De novo design protein -NX5 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design protein -NX5
To Be Published
|
|
8KC8
 
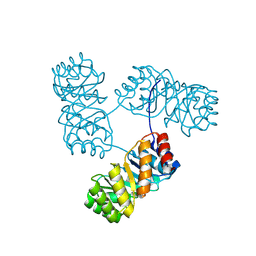 | | De novo design protein -T11 | | Descriptor: | De novo design protein -T11 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design protein -T11
To Be Published
|
|
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
5VQD
 
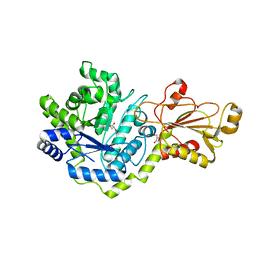 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
5KOW
 
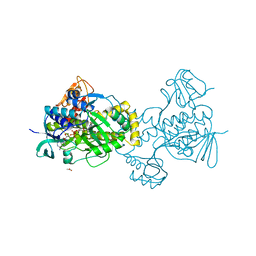 | | Structure of rifampicin monooxygenase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | Authors: | Tanner, J.J, Liu, L.-K. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
