8IGB
 
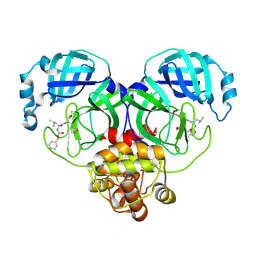 | |
2J11
 
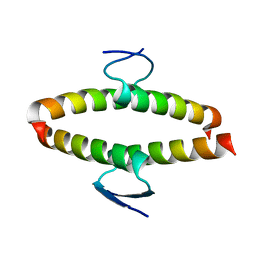 | | p53 tetramerization domain mutant Y327S T329G Q331G | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
5FMU
 
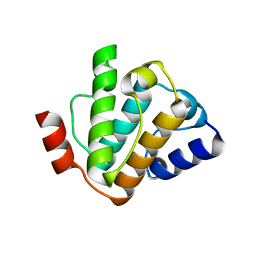 | | MmIFT54 CH-domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TRAF3-INTERACTING PROTEIN 1 | | Authors: | Weber, K, Lorentzen, E. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Intraflagellar Transport Proteins 172, 80, 57, 54, 38, and 20 Form a Stable Tubulin-Binding Ift-B2 Complex.
Embo J., 35, 2016
|
|
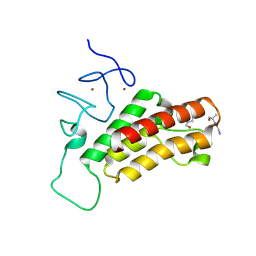
3O36
 
 | |
8J7E
 
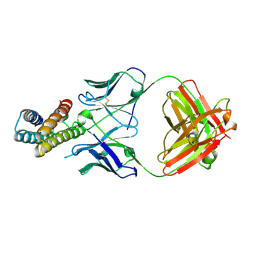 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
5FZV
 
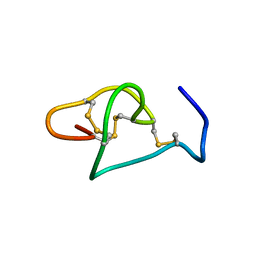 | |
5FZX
 
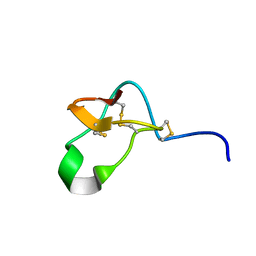 | |
5FZW
 
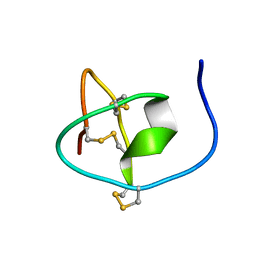 | |
7CNU
 
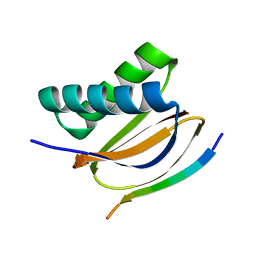 | | Crystal structure of DLC2 in complex with BMF peptide | | Descriptor: | Bcl-2-modifying factor, Dynein light chain 2, cytoplasmic | | Authors: | Wen, Y, Shao, Y. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-canonical phosphorylation of Bmf by p38 MAPK promotes its apoptotic activity in anoikis.
Cell Death Differ., 29, 2022
|
|
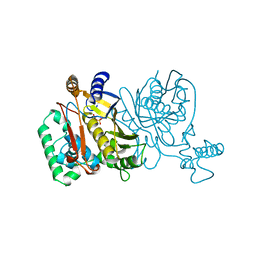
1JMF
 
 | |
5FM9
 
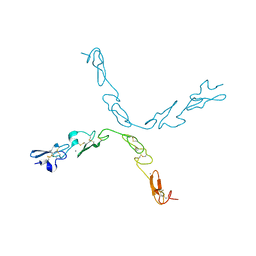 | | human Notch 1, EGF 4-7 | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Weisshuhn, P.C, Sheppard, D, Taylor, P, Whiteman, P, Lea, S.M, Handford, P.A, Redfield, C. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Non-Linear and Flexible Regions of the Human Notch1 Extracellular Domain Revealed by High-Resolution Structural Studies.
Structure, 24, 2016
|
|
1ORI
 
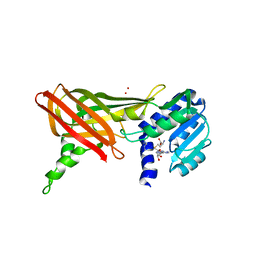 | | Structure of the predominant protein arginine methyltransferase PRMT1 | | Descriptor: | Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of its Binding to Substrate Peptides
Structure, 11, 2003
|
|
1JMH
 
 | |
1JMI
 
 | |
5GSQ
 
 | | Crystal structure of IgG Fc with a homogeneous glycoform and Antibody-Dependent Cellular Cytotoxicity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, C.-L, Hsu, J.-C, Lin, C.-W, Wu, C.-Y, Wong, C.-H, Ma, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a Homogeneous IgG-Fc Glycoform with the N-Glycan Designed to Maximize the Antibody Dependent Cellular Cytotoxicity
ACS Chem. Biol., 12, 2017
|
|
6Y6B
 
 | | Crystal structure of human 14-3-3 gamma in complex with CaMKK2 14-3-3 binding motif Ser100 and 16-OMe-Fusicoccin H | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[[(1~{S},3~{R},6~{S},7~{S},8~{R},9~{R},10~{R},11~{R},14~{S})-14-(me thoxymethyl)-3,10-dimethyl-9-oxidanyl-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradecanyl]oxy]oxane-3,4,5-triol, 14-3-3 protein gamma, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Lentini Santo, D, Obsilova, V, Obsil, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Acs Chem.Biol., 15, 2020
|
|
1KBA
 
 | |
5GG7
 
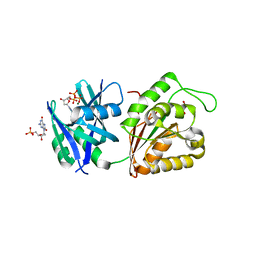 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP, 8-oxo-dGMP and pyrophosphate (I) | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGC
 
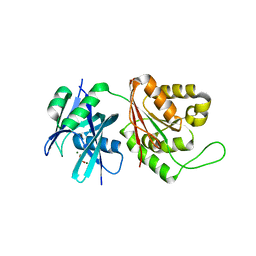 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with phosphate and magnesium ions (excess magnesium, I) | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGX
 
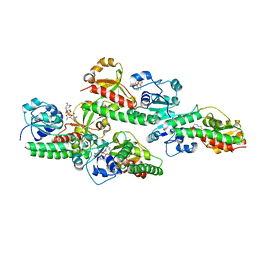 | |
5GHI
 
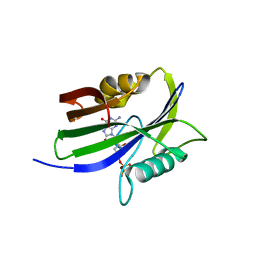 | | Crystal structure of human MTH1(G2K mutant) in complex with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHO
 
 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
6FLG
 
 | |
1PGJ
 
 | | X-RAY STRUCTURE OF 6-PHOSPHOGLUCONATE DEHYDROGENASE FROM THE PROTOZOAN PARASITE T. BRUCEI | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, SULFATE ION | | Authors: | Dohnalek, J, Phillips, C, Gover, S, Barrett, M.P, Adams, M.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A 2.8 A resolution structure of 6-phosphogluconate dehydrogenase from the protozoan parasite Trypanosoma brucei: comparison with the sheep enzyme accounts for differences in activity with coenzyme and substrate analogues.
J.Mol.Biol., 282, 1998
|
|
2YC4
 
 | | Intraflagellar Transport Complex 25-27 from Chlamydomonas | | Descriptor: | CALCIUM ION, CHLORIDE ION, INTRAFLAGELLAR TRANSPORT PROTEIN 25, ... | | Authors: | Bhogaraju, S, Taschner, M, Lorentzen, E. | | Deposit date: | 2011-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Intraflagellar Transport Complex 25/27.
Embo J., 30, 2011
|
|
