2GFT
 
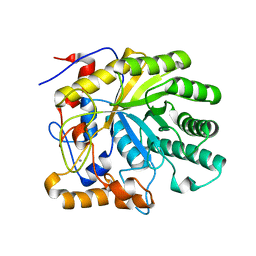 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
2GGL
 
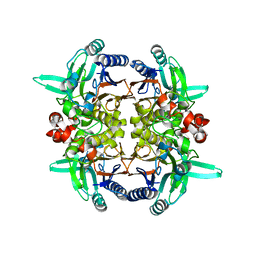 | |
2GAC
 
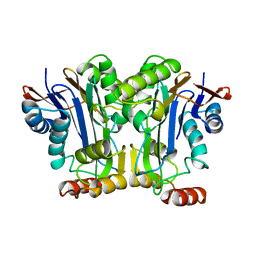 | |
5A0A
 
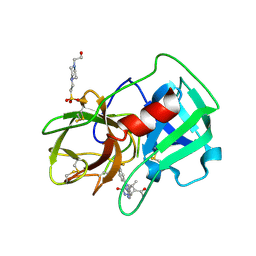 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-[(6R)-5-ethanoyl-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidin-6-yl]pyridine-2-carbonitrile, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5AE5
 
 | | Structures of inactive and activated DntR provide conclusive evidence for the mechanism of action of LysR transcription factors | | Descriptor: | GLYCEROL, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Lerche, M, Dian, C, Round, A, Lonneborg, R, Brzezinski, P, Leonard, G.A. | | Deposit date: | 2015-08-25 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | The Solution Configurations of Inactive and Activated Dntr Have Implications for the Sliding Dimer Mechanism of Lysr Transcription Factors.
Sci.Rep., 6, 2016
|
|
5AGA
 
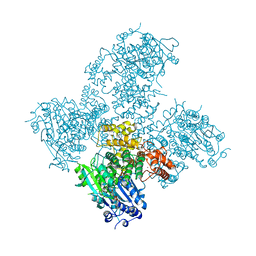 | | Crystal structure of the Helicase domain of human DNA polymerase theta in complex with AMPPNP | | Descriptor: | CITRATE ANION, DNA POLYMERASE THETA, MAGNESIUM ION, ... | | Authors: | Newman, J.A, Cooper, C.D.O, Aitkenhead, H, Pinkas, D.M, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-25 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway.
Structure, 23, 2015
|
|
2GID
 
 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
1AOS
 
 | |
5A0C
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | (6S)-6-(4-cyano-2-methylsulfonyl-phenyl)-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-5-carbonitrile, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5M26
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with methylhydroquinone | | Descriptor: | 2-methylbenzene-1,4-diol, FE (III) ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5A09
 
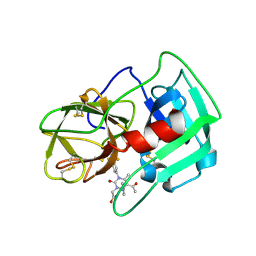 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-[(4R)-4-(4-cyanophenyl)-5-ethanoyl-6-methyl-2-oxidanylidene-1-[3-(trifluoromethyl)phenyl]-4H-pyrimidin-3-yl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUTROPHIL ELASTASE, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
6M4T
 
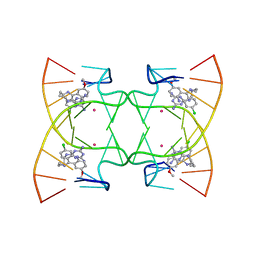 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*(UD)P*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), N4-[4-[(6-chloranyl-2-methoxy-acridin-9-yl)amino]butyl]-1,3,5-triazine-2,4,6-triamine | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B, Wu, P.C. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
5AD1
 
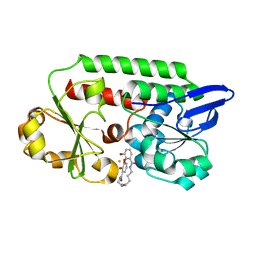 | | A complex of the synthetic siderophore analogue Fe(III)-8-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-OCTANE-1,8-DIYLBIS(2,3-DIHYDROXYBENZAMIDE) | | Authors: | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
6YND
 
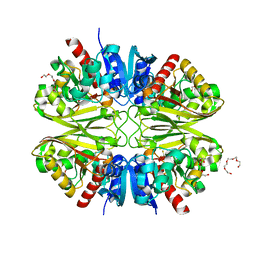 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 1 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6M5J
 
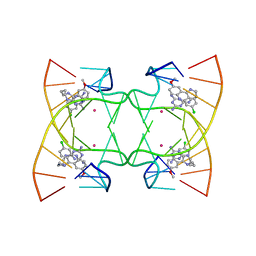 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA/TTCTGCAGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*TP*TP*CP*TP*GP*CP*AP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
4XFP
 
 | | Crystal Structure of Highly Active Mutant of Bacillus sp. TB-90 Urate Oxidase | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Hibi, T, Hayashi, Y, Kawamura, A, Itoh, T. | | Deposit date: | 2014-12-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Glycine Substitution of Surface Proline 287 Involves Entropic Enhancement of Bacillus sp. TB-90 Uricase Activity
To be published
|
|
5I79
 
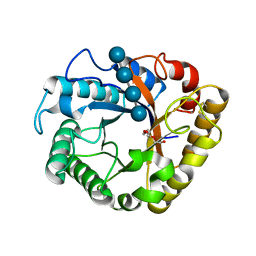 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
1OOZ
 
 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7YUV
 
 | | MtaLon-ADP for the spiral oligomers of tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
5A6X
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with alpha-methyl-fucoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-L-fucopyranoside | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
7YUM
 
 | | MtaLon-Apo for the spiral oligomers of tetramer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
5AJC
 
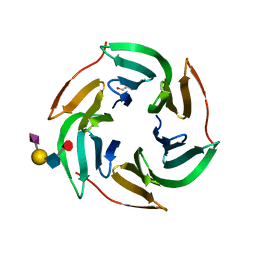 | | X-ray structure of RSL lectin in complex with sialyl lewis X tetrasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Topin, J, Arnaud, J, Varrot, A, Imberty, A. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hidden Conformation of Lewis X, a Human Histo-Blood Group Antigen, is a Determinant for Recognition by Pathogen Lectins
Acs Chem.Biol., 11, 2016
|
|
6YNF
 
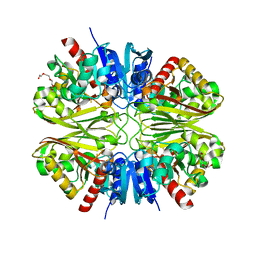 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 3 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
1OPE
 
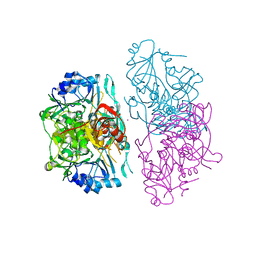 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-05 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5IOS
 
 | |
