3RFT
 
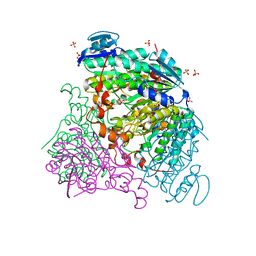 | |
3R1N
 
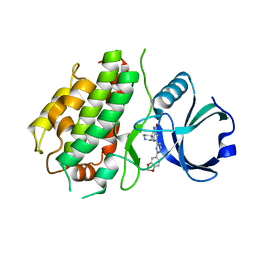 | | MK3 kinase bound to Compound 5b | | Descriptor: | 2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one, MAP kinase-activated protein kinase 3 | | Authors: | Oubrie, A, Kazemier, B. | | Deposit date: | 2011-03-11 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SAD
 
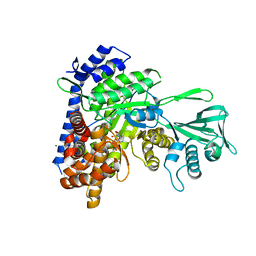 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 4-(2-mehtylphenyl)-2,4-dioxobutanoic acid inhibitor | | Descriptor: | 4-(2-methylphenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-02 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
3S04
 
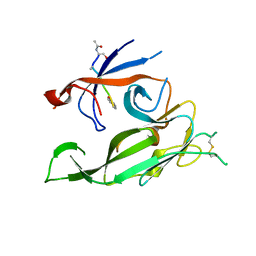 | |
3RVI
 
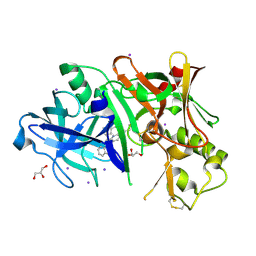 | |
3P9C
 
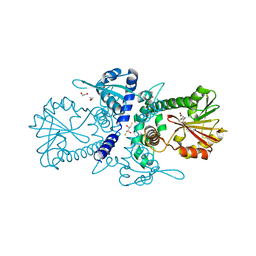 | | Crystal structure of perennial ryegrass LpOMT1 bound to SAH | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
3UBX
 
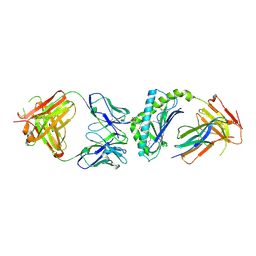 | | Crystal structure of the mouse CD1d-C20:2-aGalCer-L363 mAb Fab complex | | Descriptor: | (11Z,14Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-10-25 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the recognition of C20:2-alpha GalCer by the invariant natural killer T cell receptor-like antibody L363.
J.Biol.Chem., 287, 2012
|
|
3OVX
 
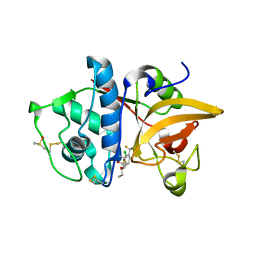 | |
4F08
 
 | |
4F2Y
 
 | | Structure of 3'-Fluoro Cyclohexenyl Nucleic Acid Decamer | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(XTF)P*AP*CP*GP*C)-3', MAGNESIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Synthesis and Antisense Properties of Fluoro Cyclohexenyl Nucleic Acid (F-CeNA), a Nuclease Stable Mimic of 2'-Fluoro RNA.
J.Org.Chem., 77, 2012
|
|
3UUB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEY
 
 | |
4F2X
 
 | | Structure of 3'-Fluoro Cyclohexenyl Nucleic Acid Heptamer | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTF)P*GP*CP*G)-3', COBALT (III) ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Antisense Properties of Fluoro Cyclohexenyl Nucleic Acid (F-CeNA), a Nuclease Stable Mimic of 2'-Fluoro RNA.
J.Org.Chem., 77, 2012
|
|
3P9J
 
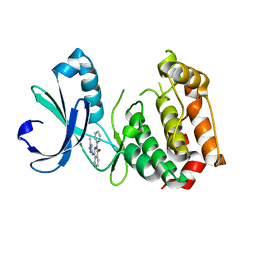 | | Aurora A kinase domain with phthalazinone pyrazole inhibitor | | Descriptor: | 4-[(5-methyl-1H-pyrazol-3-yl)amino]-2-phenylphthalazin-1(2H)-one, Serine/threonine-protein kinase 6 | | Authors: | Kairies, N.A, Oliveira, T, Engh, R.A. | | Deposit date: | 2010-10-17 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phthalazinone pyrazoles as potent, selective, and orally bioavailable inhibitors of aurora-a kinase.
J.Med.Chem., 54, 2011
|
|
3OVZ
 
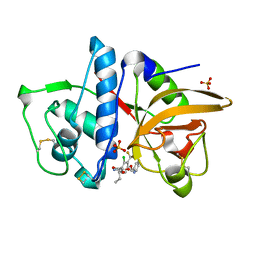 | | Cathepsin K in complex with a covalent inhibitor with a ketoamide warhead | | Descriptor: | Cathepsin K, N-[(1S)-3-amino-1-ethyl-2,3-dioxopropyl]-2-chloro-4-(pyridin-2-ylmethoxy)-3-(trifluoromethyl)benzamide, SULFATE ION | | Authors: | Fradera, X, van Zeeland, M, Uitdehaag, J.C.M. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Trifluoromethylphenyl as P2 for ketoamide-based cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4EYZ
 
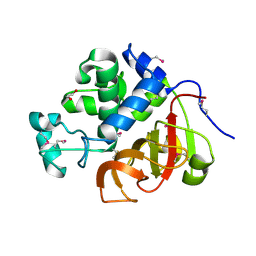 | | Crystal structure of an uncommon cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | Descriptor: | 1,2-ETHANEDIOL, Cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | Authors: | Frolow, F, Voronov-Goldman, M, Bayer, E, Lamed, R. | | Deposit date: | 2012-05-02 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Crystal Structure of an Uncommon Cellulosome-Related Protein Module from Ruminococcus flavefaciens That Resembles Papain-Like Cysteine Peptidases.
Plos One, 8, 2013
|
|
4FK9
 
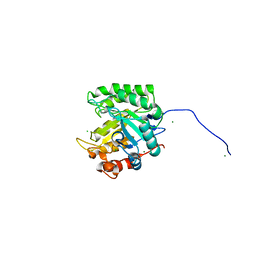 | |
4M62
 
 | |
3SBO
 
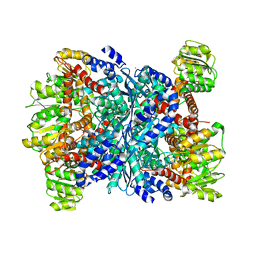 | | Structure of E.coli GDH from native source | | Descriptor: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | Authors: | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | Deposit date: | 2011-06-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3OA5
 
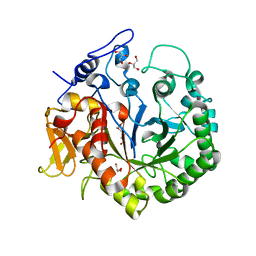 | | The structure of chi1, a chitinase from Yersinia entomophaga | | Descriptor: | Chi1, GLYCEROL, NONAETHYLENE GLYCOL | | Authors: | Busby, J.N, Lott, J.S, Hurst, M.R.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of Chi1 Chitinase from Yen-Tc: the multisubunit insecticidal ABC toxin complex of Yersinia entomophaga
J.Mol.Biol., 415, 2012
|
|
4LOI
 
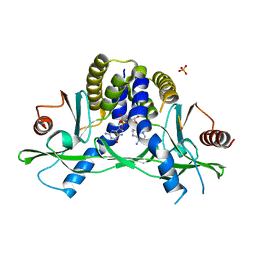 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(2',5')p] | | Descriptor: | 2-amino-9-[(1R,3R,6R,8R,9R,11S,14R,16R,17R,18R)-16-(6-amino-9H-purin-9-yl)-3,11,17,18-tetrahydroxy-3,11-dioxido-2,4,7,10,12,15-hexaoxa-3,11-diphosphatricyclo[12.2.1.1~6,9~]octadec-8-yl]-1,9-dihydro-6H-purin-6-one, PHOSPHATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOK
 
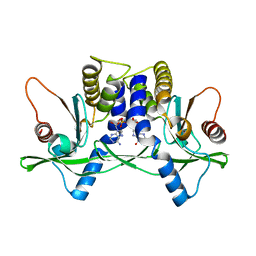 | | Crystal structure of mSting in complex with c[G(3',5')pA(3',5')p] | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
3WOW
 
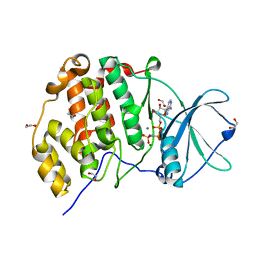 | | Crystal structure of human CK2a with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Kinoshita, T, Nakaniwa, T, Sekiguchi, Y, Sogabe, Y, Sakurai, A, Nakamura, S, Nakanishi, I, Shimada, K, Tanaka, M. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A hydrophobic residue divergence of CK2a contribute to a species-dependent variation for apigenin binding mode but not for an ATP analogue
To be Published
|
|
5SXL
 
 | |
4K99
 
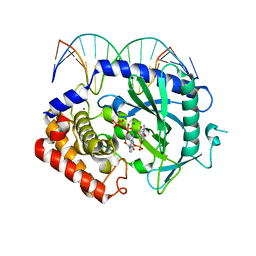 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppdG(2 ,5 )pdG | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 3'-deoxy-guanosine 5'-monophosphate, Cyclic GMP-AMP synthase, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
