3RPI
 
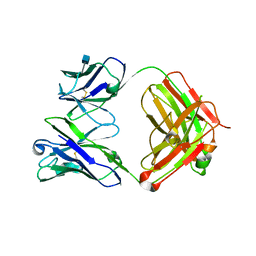 | | Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from highly potent anti-HIV neutralizing antibody, Light chain from highly potent anti-HIV neutralizing antibody | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Science, 333, 2011
|
|
3U6S
 
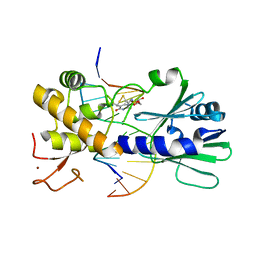 | | MutM set 1 TpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U80
 
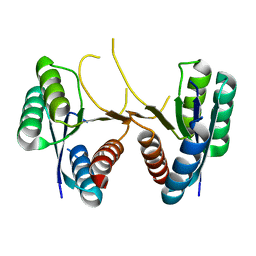 | | 1.60 Angstrom Resolution Crystal Structure of a 3-Dehydroquinate Dehydratase-like Protein from Bifidobacterium longum | | Descriptor: | 3-dehydroquinate dehydratase, type II | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type II dehydroquinate dehydratase-like protein from Bifidobacterium longum.
J.Struct.Funct.Genom., 14, 2013
|
|
3TJD
 
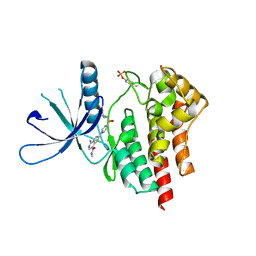 | | co-crystal structure of Jak2 with thienopyridine 19 | | Descriptor: | 4-amino-2-[4-(tert-butylsulfamoyl)phenyl]-N-methylthieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Huang, X. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
3QIO
 
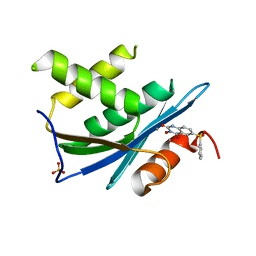 | | Crystal Structure of HIV-1 RNase H with engineered E. coli loop and N-hydroxy quinazolinedione inhibitor | | Descriptor: | 3-hydroxy-6-(phenylsulfonyl)quinazoline-2,4(1H,3H)-dione, Gag-Pol polyprotein,Ribonuclease HI,Gag-Pol polyprotein, MANGANESE (II) ION, ... | | Authors: | Lansdon, E.B, Liu, Q. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4011 Å) | | Cite: | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3QXT
 
 | |
3QXU
 
 | |
3QXE
 
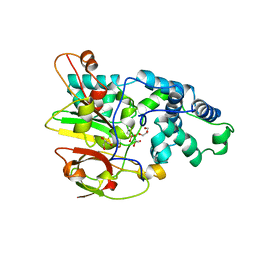 | | Crystal Structure of Co-type Nitrile Hydratase from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QUZ
 
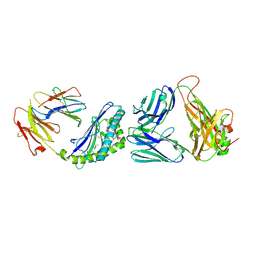 | | Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
3SW0
 
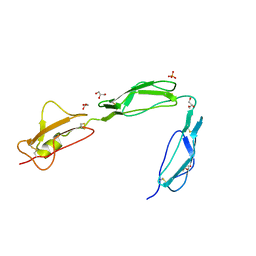 | | Structure of the C-terminal region (modules 18-20) of complement regulator Factor H | | Descriptor: | Complement factor H, GLYCEROL, PHOSPHATE ION | | Authors: | Morgan, H.P, Guariento, M, Schmidt, C.Q, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the C-Terminal Region (Modules 18-20) of Complement Regulator Factor H (FH).
Plos One, 7, 2012
|
|
3SX4
 
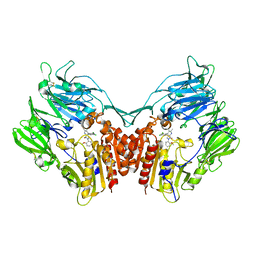 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SZV
 
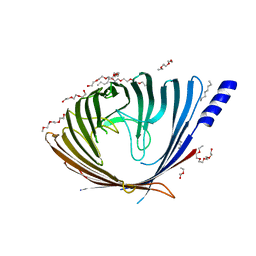 | |
3U6D
 
 | | MutM set 1 GpGo | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*(8OG)P*GP*AP*(TX)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U65
 
 | | The Crystal Structure of Tat-P(T) (Tp0957) | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, Tp33 protein | | Authors: | Brautigam, C.A, Tomchick, D.R, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3TNY
 
 | | Structure of YfiY from Bacillus cereus bound to the siderophore iron (III) schizokinen | | Descriptor: | YfiY (ABC transport system substrate-binding protein), [2-(hydroxy-kappaO)-4-[(3-{(hydroxy-kappaO)[1-(hydroxy-kappaO)ethenyl]amino}propyl)amino]-2-{2-[(3-{(hydroxy-kappaO)[1- (hydroxy-kappaO)ethenyl]amino}propyl)amino]-2-oxoethyl}-4-oxobutanoato(6-)-kappaO]iron | | Authors: | Clifton, M.C. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Parsing the functional specificity of Siderocalin / Lipocalin 2 / NGAL for siderophores and related small-molecule ligands
To be Published
|
|
3TO7
 
 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3U6C
 
 | | MutM set 1 ApGo | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*GP*AP*TP*CP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*CP*AP*(8OG)P*GP*AP*(TX)P*CP*TP*AP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3TLS
 
 | | The GLIC pentameric Ligand-Gated Ion Channel E19'P mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3QQ8
 
 | |
3Q61
 
 | | 3'-Fluoro Hexitol Nucleic Acid DNA Structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F3H)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.R, Prakash, T.P, Siwkowski, A, Berdeja, A, Yu, J, Pallan, P.S, Watt, A.T, Gaus, H, Bhat, B, Egli, M, Swayze, E.E. | | Deposit date: | 2010-12-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3'-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides.
J.Am.Chem.Soc., 133, 2011
|
|
3RQC
 
 | | Crystal structure of the catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrott, N.L, Crennell, S.J, Hough, D.W, Danson, M.J, van den Elsen, J.M.H. | | Deposit date: | 2011-04-28 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly.
Febs J., 279, 2012
|
|
3U6P
 
 | | MutM set 1 GpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3TOB
 
 | |
3TY0
 
 | | Structure of PPARgamma ligand binding domain in complex with (R)-5-(3-((3-(6-methoxybenzo[d]isoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)methyl)phenyl)-5-methyloxazolidine-2,4-dione | | Descriptor: | (5R)-5-(3-{[3-(6-methoxy-1,2-benzoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl]methyl}phenyl)-5-methyl-1,3-oxazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Soisson, S.M, Meinke, P.M, McKeever, B, Liu, W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Benzimidazolones: a new class of selective peroxisome proliferator-activated receptor gamma (PPAR-gamma) modulators.
J.Med.Chem., 54, 2011
|
|
3U6L
 
 | | MutM set 2 CpGo | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
